the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Optical and impedimetric study of genetically modified cells for diclofenac sensing
Margarita Guenther
Falko Altenkirch
Kai Ostermann
Gerhard Rödel
Ingo Tobehn-Steinhäuser
Steffen Herbst
Stefan Görlandt
Gerald Gerlach
Whole-cell biosensors, based on genetically modified yeast cells, were employed to detect anthropogenic micropollutants (e.g. drugs). Specific stimuli, e.g. traces of drugs, lead to the induction of fluorescence in the respective cells. Receptors of the cells detect specific signal molecules and induce the formation of fluorescent proteins. In this work, genetically modified cells of the yeast Saccharomyces cerevisiae BY4741 were confined in a four-chamber microfluidic cell, providing an optical monitoring of the cell behaviour and their supply with the nutrients. The measurements of the time-dependent fluorescence intensity were performed with different concentrations of the drug diclofenac, and the sensitivity of yeast cells to diclofenac was demonstrated. Cell viability was monitored by simultaneous impedance recording.
- Article
(1062 KB) - Full-text XML
-
Supplement
(668 KB) - BibTeX
- EndNote
Anthropogenic micropollutants such as drugs are often barely removed in water treatment plants, eventually entering the water cycle. Pharmaceutical residues in natural and treated waters have ecotoxicological effects on the aquatic wildlife and can lead to organ damage and general impairment in animals. The anti-inflammatory drug diclofenac is one of the most prescribed and most frequently detected compounds in wastewaters and the environment. Low concentrations (1 µg L−1) of diclofenac can lead to death of some fish species due to liver and kidney failure (Schwaiger et al., 2004).
In practice, there are very accurate and reliable analytical methods available for the detection of diclofenac in surface water, e.g. liquid chromatography–tandem mass spectrometry (Hernando et al., 2006; Ferrer-Aguirre et al., 2016), high-resolution mass spectrometry with liquid chromatography (Richardson and Ternes, 2014), gas chromatography–mass spectrometry (Deng et al., 2003), or ultra performance liquid chromatography (Nováková et al., 2006; Petrie et al., 2015). However, they require time-consuming and costly sampling and subsequent analysis in laboratory environment and necessitate special laboratories with appropriate staff and logistical efforts. Therefore, small, sensitive, robust, and online capable biosensors are needed for on-site applications (Steinke et al., 2018; Rau et al., 2014).
In this context, whole-cell biosensors, based on genetically modified yeast cells, which produce a fluorescent protein upon sensing diclofenac, are of great interest because of their low-cost mass production and cost-effective instrumentation, and the possibility of integrating them into multi-array or microprocessor-controlled diagnostic tools. Multisensors, based on living cells as biological transducers, could contribute to an inline-capable wastewater analysis approach. Receptors of the cells detect specific signal molecules, and directly or upon activation of a signal cascade induce the transcription of a reporter gene, leading to the production of a fluorescent protein (Adeniran et al., 2015; Schuller et al., 2017). If specific receptors for the substance to be detected are unknown, compound-responsive promoters can be used. The possible cross-sensitivity mediated by different transcription factors (TFs) could be lowered or eliminated by incorporating additional signal-specific TF binding sites that enhance the desired signal (Schuller et al., 2017).
The diclofenac-responsive promoter of the Saccharomyces (S.) cerevisiae PDR5 gene is involved in the pleiotropic drug resistance (PDR), a mechanism that enables cells to become resistant to different cytotoxic compounds. In yeast, diclofenac targets are the mitochondrial respiratory chain subunits Rip1p of complex III and Cox9p of complex IV (Van Leeuwen et al., 2011a). Involved in diclofenac resistance in S. cerevisiae are cell wall signaling via the protein kinase C (PKC) pathway, altered zinc homeostasis, and the induction of PDR (Van Leeuwen et al., 2011b). Diclofenac induces transcription of the PDR5 gene encoding a plasma membrane ATP-binding cassette (ABC) transporter that acts as the main contributor in this PDR response (Van Leeuwen et al., 2011b). Hence, the diclofenac-responsive PDR5 promoter is an appropriate candidate for a promoter engineering approach (Schuller et al., 2017).
For quantitative determination of the reporter protein, a real-time quantification of the fraction of viable cells in the biosensor is necessary (Posseckardt et al., 2018), e.g. by fluorescence or by impedance spectroscopy or by a combination of both methods (Günther et al., 2017; Schröder et al., 2017; Tobehn-Steinhäuser et al., 2018).
In this work, a novel whole-cell biosensor is presented, in which a perfusion-designed microfluidic system is combined with an effective and long-term-stable packaging enabling the simultaneous read-out of optical and impedimetric signals. The genetically modified yeast cells are completely enclosed in this packaging; the medium continuously flows through the channels and chambers with living cells. To create a more realistic cell environment, nutrients and other important media factors were continuously supplied by fresh media and removal of waste products. The analysis of the concentration-dependent optical signal was performed during long-term measurements with diclofenac in the measurement medium. Cell metabolic activity was monitored by simultaneous impedance recording.
2.1 Chemicals
The following media were used for the culturing and suspending yeast cells.
-
Minimal medium (MM): 20 g L−1 glucose, 5 g L−1 ammonium sulfate, 2.5 g L−1 yeast nitrogen base without amino acids, 60 mg L−1 L-histidin, 80 mg L−1 L-leucine, 20 mg L−1 L-methionin (Carl Roth GmbH, Germany).
-
Synthetic defined medium (SD) (Formedium, UK): 2 % glucose, 0.17 % yeast nitrogen base without ammonium sulfate and without amino acids, 0.5 % ammonium sulfate.
-
Synthetic complete medium (SC): 6.9 g L−1 yeast nitrogen base (of which 5 g L−1 is (NH4)2SO4; Formedium, UK), 20 g L−1 d(+)-glucose (Carl Roth GmbH, Germany), and 2.002 g L−1 Kaiser mix SC complete (Formedium, UK).
-
Yeast extract peptone dextrose (YPD): 2 % glucose, 2 % bacto-peptone, 1 % yeast extract.
-
Phosphate-buffered saline (PBS): 137 mM NaCl, 2.7 mM KCl, 10.0 mM Na2HPO4, 1.76 mM KH2PO4, pH 7.4 (Sigma-Aldrich Chemie GmbH, Germany).
-
Buffer solution: 0.5 M NaCl, 0.5 M imidazole, 20 mM TRIS.
Potassium chloride (KCl) was purchased from Merck (Germany) and diclofenac sodium salt (D6899) from Sigma-Aldrich Chemie GmbH (Germany). Low-melting agarose (Plaque agarose, 840101) was obtained from Biozym Scientific GmbH (Germany).
2.2 Cells and cell culture
In this study, cells of the Saccharomyces cerevisiae (S.cerevisiae) strain BY4741 were used. In order to verify the sensitivity of the photonic transducer, the photodiode current was measured in dependence on the concentration of isolated enhanced green fluorescent protein (eGFP) as well as of S.cerevisiae BY4741-p426GPD-eGFP cells that constitutively express eGFP (Mumberg et al., 1995) (for details, see the data in Sect. S1 and Figs. S2, S3, and S4 of the Supplement). However, in the whole-cell sensor device, S. cerevisiae BY4741 p426PDR5-tGFP cells were used because of the faster folding of tGFP compared to eGFP (Evdokimov et al., 2006). Cells transformed with the multicopy plasmid p426PDR5-tGFP, applying the 5' untranslated region (UTR) of the PDR5 gene, show a diclofenac-dependent expression of the turbo green fluorescent protein (tGFP) (Schuller et al., 2017; Van Leeuwen et al., 2011b).
Preparation of S. cerevisiae BY4741 p426PDR5-tGFP cells
Yeast cells of strain BY4741 were transformed with 1 µg plasmid p426PDR5-tGFP, utilizing Zymo Research Yeast transformation kit II according to the manufacturer's instructions. Transformed cells from overnight cultures (minimal medium in conical flasks, 30 ∘C, 180 rpm) were centrifuged (3000×g, 1 min, 20 ∘C), washed once in minimal medium, and resuspended in minimal medium. The optical density at λ=600 nm of an appropriate diluted sample was determined with a VWR V-1200 spectrophotometer. For subsequent experiments, the required number of cells were centrifuged down and resuspended in the corresponding volume of minimal medium.
The cell numbers were determined by measuring the optical density of the cell suspension at 600 nm (OD600). The relationship between the cell number and OD600 was linear only for OD600≤0.3. Therefore, the cell suspension was diluted in order to obtain OD600 between 0.1 and 0.2. Afterwards, the cell numbers were calculated with an OD600=1 corresponding to 107 cells mL−1. Additionally, the cell numbers were controlled by means of the transmission-type inverted microscope Bresser-Science IVM-401 with the Neubauer improved cell counting chamber.
To adjust the optical density of OD600=2, the cell number was determined as stated above, pelleted by centrifugation, washed, and resuspended in double-concentrated MM. In order to immobilize cells, 40 µL of the cell suspension with OD600=2 in double-concentrated MM was mixed with 40 µL of 1.5 % low-melting agarose in distilled water at 50 ∘C, resulting in OD600=1 and a 0.75 % agarose in single MM. Afterwards, each of the lower chambers (a measuring chamber and a reference chamber) of the four-chamber microfluidic cell, placed on the glass substrate, was filled with 40 µL of the cell suspension–gel mixture and covered with a porous polycarbonate (PC) membrane (Fig. 1). After the installation of the upper part of the microfluidic cell, two chambers of the upper part were connected to a pump. The measurement started after solidification of the cell suspension–gel mixture by cooling down to 21 ∘C. During the measurements, the upper microfluidic chamber of the measuring channel was overflowed with diclofenac (DCF) containing MM, whereas MM without diclofenac was pumped through the reference channel.
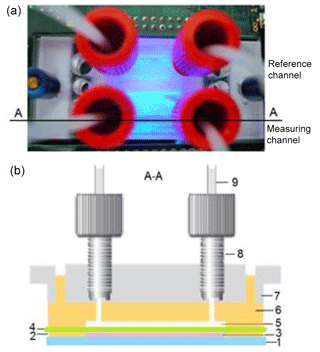
Figure 1Design of the four-chamber microfluidic cell with measuring and reference channels: top view (a) and cross section through the measuring and reference channel, respectively (b). (1) Glass substrate with coated ITO interdigitated electrode arrays, (2) PDMS foil (21.5 mm × 21.5 mm × 0.5 mm) with two openings (15 mm × 5 mm), which form the lower chambers, (3) yeast cells immobilized in gel in the lower chamber, (4) porous polycarbonate membrane, (5) upper chamber in PDMS, (6) upper PDMS part with fluidic micro-channels, (7) 22x22 MicCell PC rohling system (GeSiM mbH) with microfluidic connectors, (8) fitting UNF 1/4-28, and (9) Tygon®tube OD with a ferrule.
2.3 Design of the microfluidic cell
The living cells were immobilized on the glass substrate in a four-chamber microfluidic cell, allowing the independent measurements for the measuring and reference channels (Fig. 1). The optical transmitter–receiver module was installed below the glass substrate. For the monitoring of the cell behaviour, a microfluidic cell (made from polydimethylsiloxane (PDMS), Sylgard 184, Dow-Corning Inc.) was used. Two lower chambers (15 mm × 5 mm × 0.5 mm) of the four-chamber microfluidic cell, placed on the glass substrate, were filled with 40 µL of the cell suspension–gel mixture and covered with a porous polycarbonate membrane (Unique-Mem® Track-Etched Membrane, thickness 20 µm, pore size 1 µm, Oxyphen AG, Wetzikon, Switzerland). Afterwards, the upper part of the microfluidic cell made using a 22×22 MicCell polycarbonate rohling system (GeSiM mbH, Radeberg, Germany) (Gast et al., 2006) was installed. Two chambers of the upper part were connected to a pump and served for the nutrient or measuring solution supply with a rate of 20 µL min−1. The yeast cells (cell size ca. 5 µm) were completely enclosed in this packaging, had no contact with the environment, and were supplied with the nutrients. The packaging enabled a simultaneous monitoring of optical and impedimetric signals by using glass substrates with coated transparent ITO (indium tin oxide) interdigitated electrode arrays (see Fig. S5).
2.4 Optical read-out
The optical signal was detected using a photonic transducer SFD100 (OPTO-MST Sensoren und Systeme GmbH, Erfurt, Germany), based on a photodiode OC808 (effective active area 1 cm2). The transducer was integrated in an optical module (CiS Forschungsinstitut für Mikrosensorik GmbH, Erfurt, Germany) comprising two blue LED light sources (470 nm, one for each channel) and the corresponding excitation–emission filter set for the green fluorescent (GFP) proteins (Tobehn-Steinhäuser et al., 2018). The photodiode current was measured by means of a current-voltage converter, based on an operational amplifier OP177, and a LabJack U6-PRO card.
The LED light sources and the corresponding excitation–emission filter set were chosen in accordance with the excitation–emission spectral ranges of the green fluorescent proteins. The fluorescence of tGFP (absorption maximum at 482 nm, emission maximum at 503 nm, Evdokimov et al., 2006; Tsien, 1998; Mujiono et al., 2015) was excited by a blue LED light source (470 nm, Fig. S1) and detected by an OC808 photodiode with an OG515 long-pass emission filter.
The LED light source was supplied with a constant current of 8 mA at 26 mV. The temperature near the sample was measured using a temperature sensor LM61CIZ (Texas Instruments Inc.). During the operation of the LED light source for 0.5 min, the temperature increase in the sample plane was less than 0.5 K. The experiments were carried out under clean-room conditions at a constant temperature of 21 ∘C.
All experiments were performed as biological triplicates. Afterwards, the mean values and standard deviation (SD) were calculated. The concentration dependence of the optical signal was fitted with OriginPro (Version 9.1G) using the Fitting analysis.
Data analysis
A four-parameter logistic function (Eq. 1) was used for a regression analysis of the sigmoidal dose-response curve:
where x is the concentration of diclofenac and y is the optical signal. The range between the lower (A1) and upper (A2) asymptotes is defined as the dynamic signal range, whereas only the 10 %–90 % block of this range is determined as a working range. The inflection point (x0) represents the analyte concentration which corresponds to an increase of 50 % in the dynamic signal range (effective concentration, EC50). The slope of the tangent in the inflection point is given by the parameter p.
According to the rules of the German standard DIN 32645:2008-11 (German standard DIN 32645:2008-11, 2008), the limit of detection xLoD was calculated as
where sL is the standard deviation of the signal without the analyte (i.e. of the blank measurements) and p is the slope of the calibration curve. The factor k=3 was used in compliance with the IUPAC rules (Inczedy et al., 1998; IUPAC, 2014).
The time-dependent increase in the optical signal in the presence of diclofenac was fitted using the modified Gompertz model (Zwietering et al., 1990):
with the lag phase duration λ and the maximum specific growth rate μmax. For the optical signal, applies, where I0 is the initial value of the signal and It is the signal at time t; the asymptote is the saturation value reached with Imax, and e=exp (1). The maximum specific growth rate μmax is defined as the tangent in the inflection point of the three-phase growth curve. The lag time λ is defined as the t-axis intercept of this tangent. The inflection point (t50) represents the time of a half-maximal effect. This model is often used in biochemistry to describe the behaviour of microorganisms under different physical or chemical conditions (Arroyo-López et al., 2009; Zwietering et al., 1990).
Alternatively, in order to better fit an initial phase of the growth curve, the following two-part logistic function can be used:
2.5 Impedimetric read-out
In order to enable a combination of optical and electrical measurements, transparent ITO (indium tin oxide, 0.5 µm thick) interdigital structures for impedance spectroscopy were coated on the B270i glass wafers (wafer thickness (1.0±0.1) mm) (Fig. S5). The prepared structures were optically transparent and conductive. The living cells were immobilized on the glass substrate with two coated ITO-electrode arrays in a four-chamber microfluidic cell.
Two ITO-electrode arrays provided the independent impedance measurements for the measuring and reference channels. Each array consists of 24 fingers (with a finger length L of 11 mm). The distance S between the fingers varies between 2.5 and 133.5 µm and the width W between 5 and 186 µm (Fig. S5 and Table S1). Using a multiplexer, it was possible to connect the finger structures arbitrarily to form arrays or to operate them individually. As a result, the penetration depth d of the electric field into the medium could be varied and estimated as (Mamishev et al., 2004). The multiplexer board (CiS, Erfurt, Germany) consists of a main board and a needle adapter board. The latter was detachably connected to the main board by two ZIF flex connections. It consists of a 2×12 dual analogue multiplexer, 2×24 spring contact pins, and four micro coax connectors. The main board was connected to an impedance analyser ISX-3v2 (Sciospec GmbH, Germany). The recorded impedance spectrum of the genetically modified yeast cells at t=0 h, shown in Fig. S6, was used as a reference.
The cell constant Kcell=5.4 cm−1 was determined for interdigitated structures using a calibration medium of 0.1 M KCl solution (Fig. S6).
The sensor concept, based on living cells as biological transducers, was validated using genetically modified Saccharomyces cerevisiae cells (strain BY4741), which express the turbo green fluorescent protein (tGFP).
Transformed S. cerevisiae BY4741 p426PDR5-tGFP cells, which express tGFP under the control of the diclofenac-responsive PDR5 gene promoter, were immobilized in a 0.75 % agarose gel and confined in a microfluidic cell, installed on the glass substrate. Two lower chambers of the four-chamber microfluidic cell, placed on the glass substrate, were filled with the cell suspension–gel mixture and covered with a porous polycarbonate (PC) membrane. Afterwards, the upper part of the microfluidic cell was installed. Two chambers of the upper part were connected to a pump and served for the supply of nutrient (MM in the reference channel) and measuring solution (diclofenac containing MM in the measuring channel). The same diclofenac-sensitive cells were immobilized in the lower chambers of the measuring and reference channels. Therefore, both channels were irradiated with light of the same wavelength (470 nm). The optical measurements were performed by periodic switching between the LED light sources of the measuring and reference channels. The difference between the values of the photodiode current of these channels was taken as an optical signal.
Figure 2 shows the change in the photodiode current IPD as a function of the diclofenac concentration after a 16 h overflow with DCF containing MM. For each DCF concentration, newly cultured cells were used. The optical signal was determined using the optical module, which was located below the microfluidic cell containing the immobilized yeast cells. The drift of the photodiode current reached its minimal value of 5 nA s−1 at the excitation time of 0.5 min. Therefore, the IPD values at the excitation time te=0.5 min were used to determine the characteristic concentration–response curve of the sensor. The data clearly document that the fluorescence increases with increasing DCF concentration. Upon detecting a DCF molecule with specific receptors, the cells produce a fluorescent reporter protein. We observed that an impedance increase appeared in solution with diclofenac during long-term measurements (Fig. 3). This may be caused by increasing protein production.
Table 1Limit of detection (LoD) and working range for detection of diclofenac in comparison with literature values.
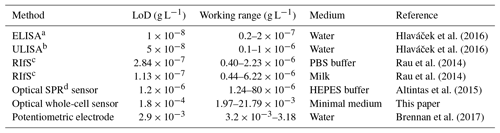
a Enzyme-linked immunosorbent assay (ELISA). b Upconversion-linked immunosorbent assay (ULISA). c Reflectometric interference spectroscopy (RIfS). d Surface plasmon resonance (SPR).
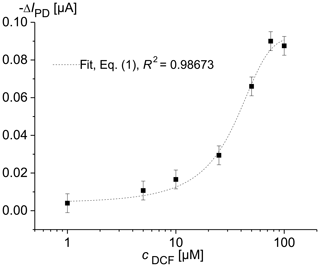
Figure 2Change in the photodiode current IPD in dependence on the concentration of diclofenac (DCF) for living S. cerevisiae BY4741 p426PDR5-tGFP cells, immobilized in 0.75 % agarose gel in the lower microfluidic chambers (7×106 cells mL−1). The upper microfluidic chamber of the measuring channel was overflowed with MM, containing DCF, at a rate of 20 µL min−1. MM without DCF was pumped through the reference channel.
The concentration–response data set was fitted using the logistic fit function of Eq. (1) with four parameters. The data set and the logistic fit function were plotted in a semilogarithmic graph, as shown in Fig. 2. The limit of detection xLoD was calculated using Eq. (2). The limit of detection (LoD) and EC50 of the calibration curve were 0.56 µM (0.18 mg L−1) and 32 µM (10.18 mg L−1), respectively. The working range was defined between 6.2 µM (1.97 mg L−1) and 68.5 µM (21.79 mg L−1). The obtained results are presented in Table 1 in comparison with literature values.
The time-dependent increase in the optical signal in the presence of diclofenac shows an initial phase in which the specific signal increase rate starts at a value of zero at the time ti=3 h and then accelerates to a maximal value µmax in a certain period of time, resulting in a lag time λ (Fig. 3). In addition, the curve contains a final phase in which the rate decreases and finally reaches zero, so that an asymptote A (see Eq. 3) is reached. The experimental curve was fitted using Eqs. (3) and (4), and the values λ=7.3 h, μmax=0.02 h−1, and t50=9.9 h were obtained for the DCF concentration of 100 µM. For DCF concentrations of 25, 50, and 75 µM, the values of 13.5, 12.1, and 10.7 h, respectively, were obtained for the time t50 of a half-maximal effect. Besides the detection after a defined time (as in Fig. 2), a kinetic analysis is also feasible.
Recent data by Schuller et al. (2017) based on fluorescence microscopy showed that S. cerevisiae BY4741 p426PDR5-tGFP cells react to diclofenac with a time delay ti that decreases with increasing DCF concentration. For DCF concentrations of 1, 10, and 100 µM, fluorescence changes were observed after 11, 6, and 3 h, respectively. The fluorescence response reached its saturation at a DCF concentration of 75 µM DCF (Schirmer et al., 2018). The smaller saturation value for 100 µM DCF is possibly due to the inhibitory effect of the high DCF concentrations on the cell growth (Van Leeuwen, 2011a). As an expression of the fluorescent proteins depends on vital cells, measuring impedance should help to expand the knowledge gained from fluorescence measurements. For example, a change in impedance might hint at different viability of the cells in the chambers.
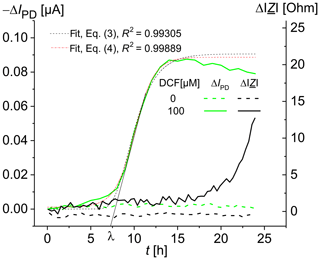
Figure 3Time-dependent change in the photodiode current IPD and the impedance modulus at 100 kHz for living S. cerevisiae BY4741 p426PDR5-tGFP cells, immobilized in 0.75 % agarose gel in the lower microfluidic chambers (7×106 cells mL−1). The upper microfluidic chambers were overflowed with MM, containing 0 µM DCF (in the reference channel) and 100 µM DCF (in the measuring channel), respectively, at a rate of 20 µL min−1.
As recently shown by means of fluorescence microscopy and flow cytometry, the fluorescence intensity of samples with S. cerevisiae BY4741 p426PDR5-tGFP cells correlates with the fraction of fluorescent cells within the cell population in dependence on the DFC concentration and the time after drug application (Schirmer et al., 2018). Therefore, in order to compare the results of different experiments, it is necessary to determine the fraction of fluorescent cells in the sample. Additionally, one has to ensure that the copy number of the PDR5-tGFP construct per cell is identical. A suitable measure to avoid copy number variation that is commonly observed in the case of multicopy plasmids (Karim et al., 2013) is the generation of yeast cells with the PDR5-tGFP module stably integrated into the genome.
For practical application of the described biosensor concept, a further improvement of the sensor performance is needed. Current approaches aim at the development of the cells towards higher sensitivity into the low nM-detection range of diclofenac by mutagenesis of the involved transcription factors and their DNA-binding sites (Schuller et al., 2017). As other substances, e.g. the antibiotic sulfamethoxazole or the endocrine disruptor 2,4-dichlorophenol, could inhibit the cell growth (Borga and Borga, 1997) and thus affect the sensor signal, the cross-sensitivity of the sensor cells has to be carefully investigated in future studies.
In this report we introduce a device of a biosensor in which immobilized yeast sensor cells are completely enclosed in a microfluidic cell with no direct contact with the environment. The packaging provided the reliable and long-term stable detection of the sensor cell-caused optical signal as well as the simultaneous read-out of the impedimetric signal for monitoring the cell viability. Solutions with different diclofenac concentrations were used to demonstrate the sensitivity of S. cerevisiae BY4741 p426PDR5-tGFP cells in the detection of the model analyte diclofenac. Ongoing experiments address the development of sensor cells with a higher sensitivity and a better signal stability, e.g. by genomic integration of the PDR5-tGFP construct, to allow the comparison of different experiments. On the hardware level, future activities will focus on the development of a small detection unit for the read-out of the fluorescence. Such sensor systems, based on the whole-cell concept, could contribute to an improved, inline-capable wastewater analysis.
The underlying measurement data are not publicly available and can be requested from the authors if required.
The supplement related to this article is available online at: https://doi.org/10.5194/jsss-8-215-2019-supplement.
Conceptualization was by GR, KO, and GG; experiments and data analysis were performed by MG; preparation of cells was done by FA; preparation of ITO-electrode arrays was done by ITS; preparation of the multiplexer board was done by SH; preparation of the optical read-out module was done by SG; writing and original draft preparation were by MG; writing review and editing were done by MG, GR, KO, and GG; visualization was by MG.
The authors declare that they have no conflict of interest.
This article is part of the special issue “Dresden Sensor Symposium 2017”. It is a result of the Dresden Sensor Symposium 2017, Dresden, Germany, 4–6 December 2017.
The authors are grateful to Christine Schirmer, Juliane Posseckardt, and Wolfgang Fichtner from the Kurt-Schwabe-Institut für Mess- und Sensortechnik e.V. Meinsberg as well as Steffen Howitz from the GeSiM mbH for fruitful discussions and the help to prepare the microfluidic cell. The financial support by the German Federal Ministry of Education and Research is kindly acknowledged (BioSAM HIGS 03WKCL01F).
This research has been supported by the German Federal Ministry of Education and Research (grant BioSAM HIGS-03WKCL01F).
This paper was edited by Fred Lisdat and reviewed by two anonymous referees.
Adeniran, A., Sherer, M., and Tyo, K. E. J.: Yeast-based biosensors: Design and applications, FEMS Yeast Res., 15, 1–15, 2015.
Altintas, Z., Guerreiro, A., Piletsky S. A., and Tothill, I. E.: NanoMIP based optical sensor for pharmaceuticals monitoring, Sensor Actuat. B-Chem., 213, 305–313, https://doi.org/10.1016/j.snb.2015.02.043, 2015.
Arroyo-López, F. N., Orli, S., Querol, A., and Barrio, E.: Effects of temperature, pH and sugar concentration on the growth parameters of Saccharomyces cerevisiae, S. kudriavzevii and their interspecific hybrid, Int. J. Food Microbiol., 131, 120–127, https://doi.org/10.1016/j.ijfoodmicro.2009.01.035, 2009.
Borga, O. and Borga, B.: Serum protein binding of nonsteroidal anti-inflammatory drugs: a comparative study, J. Pharmacokinet. Biop., 25, 63–77, 1997.
Brennan, E., Futvoie, P., Cassidy, J., and Schazmann, B.: An ionic liquid-based sensor for diclofenac determination in water, Int. J. Environ. An. Ch., 97, 588–596, https://doi.org/10.1080/03067319.2017.1333607, 2017.
Deng, A., Himmelsbach, M., Zhu, Q.-Z., Frey, S., Sengl, M., Buchberger, W., Niessner, R., and Knopp, D.: Residue analysis of the pharmaceutical diclofenac in different water types using ELISA and GC–MS, Environ. Sci. Technol., 37, 3422–3429, https://doi.org/10.1021/es0341945, 2003.
Evdokimov, A. G., Pokross, M. E., Egorov, N. S., Zaraisky, A. G., Yampolsky, I. V., Merzlyak, E. M., Shkoporov, A. N., Sander, I., Lukyanov, K. A., and Chudakov, D. M.: Structural basis for the fast maturation of Arthropoda green fluorescent protein, EMBO Rep., 7, 1006–1012, https://doi.org/10.1038/sj.embor.7400787, 2006.
Ferrer-Aguirre, A., Romero-González, R., Vidal, J. L. M., and Frenich, A. G.: Simple and quick determination of analgesics and other contaminants of emerging concern in environmental waters by on-line solid phase extraction coupled to liquid chromatography–tandem mass spectrometry, J. Chromatogr. A., 1446, 27–33, https://doi.org/10.1016/j.chroma.2016.04.009, 2016.
Gast, F.-U., Dittrich, P. S., Schwille, P., Weigel, M., Mertig, M., Opitz, J., Queitsch, U., Diez, S., Lincoln, B., Wottawah, F., Schinkinger, S., Guck, J., Käs, J., Smolinski, J., Salchert, K., Werner, C., Duschl, C., Jäger, M. S., Uhlig, K., Geggier, P., and Howitz, S.: The microscopy cell (MicCell), a versatile modular flowthrough system for cell biology, biomaterial research, and nanotechnology, Microfluid. Nanofluid., 2, 21–36, https://doi.org/10.1007/s10404-005-0047-6, 2006.
German standard DIN 32645:2008-11: Deutsches Institut für Normung e.V. (DIN) Chemische Analytik – Nachweis-, Erfassungs- und Bestimmungsgrenze unter Wiederholbedingungen – Begriffe, Verfahren, Auswertung (Chemical analysis – Decision limit, detection limit and determination limit under repeatability conditions – Terms, methods, evaluation), 2008.
Günther, M., Gerlach, G., Schirmer, C., Posseckardt, J., Fichtner, W., Mertig, M., Altenkirch, F., Ostermann, K., Schuller, A., Rödel, G., Winzer, A. T., Tobehn-Steinhäuser, I., Schröder, M., Gläser, M., Scharff, W., and Zimmermann, P.: Combined optical-impedimetric whole-cell biosensor, in: Proceedings of the 13th Sensor Symposium of Dresden, Dresden, Germany, 4–6 December 2017, AMA Association for Sensors and Measurement, 120-125, https://doi.org/10.5162/13dss2017/3.7, 2017.
Hernando, M. D., Heath, E., Petrovic, M., and Barceló, D.: Trace-level determination of pharmaceutical residues by LC-MS/MS in natural and treated waters. A pilot-survey study, Anal. Bioanal. Chem., 385, 985–991, https://doi.org/10.1007/s00216-006-0394-5, 2006.
Hlaváček, A., Farka, Z., Hübner, M., Horňáková, V., Němeček, D., Niessner, R., Skládal, P., Knopp, D., and Gorris, H. H.: Competitive upconversion-linked immunosorbent assay for the sensitive detection of diclofenac, Anal. Chem., 88, 6011–6017, https://doi.org/10.1021/acs.analchem.6b01083, 2016.
Inczedy, J., Lenguel, T., and Ure, A. M.: Compendium of analytical nomenclature. Definitive rules, in: The Orange Book, 3rd edn. Blackwell Science, Oxford, 1998.
IUPAC: Compendium of Chemical Terminology, Gold Book, Version 2.3.3, p. 839, https://doi.org/10.1351/goldbook.L03540, 2014.
Karim, A. S., Curran, K. A., and Alper, H. S.: Characterization of plasmid burden and copy number in Saccharomyces cerevisiae for optimization of metabolic engineering applications, FEMS Yeast Res., 13, 107-116, https://doi.org/10.1111/1567-1364.12016, 2013.
Mamishev, A. V., Sundara-Rajan, K., Yang, F., Du, Y., and Zahn, M.: Interdigital sensors and transducers, P. IEEE, 92, 808–845, https://doi.org/10.1109/JPROC.2004.826603, 2004
Mujiono, T., Sukekawa, Y., Nakamoto, T., Mitsuno, H., Kanzaki, R., and Misawa, N.: A cell-based odor sensing system using fluorescent technique and lock-in measurement robust against disturbance, in: Proc. IEEE Sensors 2015, 695-698, https://doi.org/10.1109/ICSENS.2015.7370351, 2015.
Mumberg, D., Muller, R., and Funk, M.: Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds, Gene, 156, 119–122, 1995.
Nováková, L., Solichová, D., and Solich, P.: Advantages of ultra performance liquid chromatography over high-performance liquid chromatography: comparison of different analytical approaches during analysis of diclofenac gel, J. Sep. Sci., 29, 2433–2443, https://doi.org/10.1002/jssc.200600147, 2006.
Petrie, B., Barden, R., and Kasprzyk-Hordern, B.: A review on emerging contaminants in wastewaters and the environment: current knowledge, understudied areas and recommendations for future monitoring, Water Res., 72, 3–27, https://doi.org/10.1016/j.watres.2014.08.053, 2015.
Posseckardt, J., Schirmer, C., Kick, A., Rebatschek, K., Lamza, T., and Mertig, M.: Monitoring of Saccharomyces cerevisiae viability by non-Faradaic impedance spectroscopy using interdigitated screen-printed platinum electrodes, Sensor Actuat. B, 255, 3417–3424, https://doi.org/10.1016/j.snb.2017.09.171, 2018.
Rau, S., Hilbig, U., and Gauglitz, G.: Label-free optical biosensor for detection and quantification of the non-steroidal anti-inflammatory drug diclofenac in milk without any sample pretreatment, Anal. Bioanal. Chem., 406, 3377–3386, https://doi.org/10.1007/s00216-014-7755-2, 2014.
Richardson, S. D. and Ternes, T. A.: Water analysis: emerging contaminants and current issues, Anal. Chem., 86, 2813–2848, https://doi.org/10.1021/ac500508t, 2014.
Schirmer, C., Posseckardt, J., Kick, A., Rebatschek, K., Fichtner, W., Ostermann, K., Schuller, A., Rödel, G., and Mertig, M.: Encapsulating genetically modified Saccharomyces cerevisiae cells in a flow-through device towards the detection of diclofenac in wastewater, J. Biotechnol., 284, 75–83, https://doi.org/10.1016/j.jbiotec.2018.08.003, 2018.
Schröder, M., Gläser, M., Schirmer, C., Posseckardt, J., Fichtner, W., Mertig, M., Tobehn-Steinhäuser, I., Herbst, S., Schädel, M., Wünscher, H., Ortlepp, T., Winzer, A., Günther, M., Altenkirch, F., Wenzel, M., Schuller, A., Rödel, G., Ostermann, K., Soltmann, U., Haufe, H., Scharff, W., and Meyer, A.: Highly integrated whole-cell sensors for the environmental and medical technology, in: Proceedings of the 13th Sensor Symposium of Dresden, Dresden, Germany, 4–6 December 2017, AMA Association for Sensors and Measurement, 148–152, https://doi.org/10.5162/13dss2017/5.2, 2017.
Schuller, A., Rödel, G., and Ostermann, K.: Tuning the sensitivity of the PDR5 promoter-based detection of diclofenac in yeast biosensors, Sensors, 17, 1506–1522, https://doi.org/10.3390/s17071506, 2017.
Schwaiger, J., Ferling, H., Mallow, U., Wintermayr, H., and Negele, R. D.: Toxic effects of the non-steroidal anti-inflammatory drug diclofenac Part I: histopathological alterations and bioaccumulation in rainbow trout, Aquat. Toxicol. 68, 141–150, https://doi.org/10.1016/j.aquatox.2004.03.014, 2004.
Steinke, N., Rio, M., Wuchrer, R., Schuster, C., Ljasenko, E., Knopp, D., Gerlach, G., and Härtling, T.: Detection of diclofenac molecules by planar and nanostructured plasmonic sensor substrates, Sensor Actuat. B-Chem., 254, 749–754, https://doi.org/10.1016/j.snb.2017.07.123, 2018.
Tobehn-Steinhäuser, I., Günther, M., Görlandt, S., Herbst, S., Wünscher, H., Ortlepp, T., and Gerlach, G.: Multisensors for whole-cell analytics, IAPGOS, 8, 39–41, https://doi.org/10.5604/01.3001.0012.0702, 2018.
Tsien, R.: The green fluorescent protein, Annu. Rev. Biochem., 67, 509–544, 1998.
Van Leeuwen, J. S., Orij, R., Luttik, M. A. H., Smits, G. J., Vermeulen, N. P. E., and Vos, J. C.: Subunits Rip1p and Cox9p of the respiratory chain contribute to diclofenac-induced mitochondrial dysfunction, Microbiology, 157, 685–694, 2011a.
Van Leeuwen, J. S., Vermeulen, N. P. E., and Vos, J. C.: Involvement of the pleiotropic drug resistance response, protein kinase C signaling, and altered zinc homeostasis in resistance of Saccharomyces cerevisiae to diclofenac, Appl. Environ. Microbiol., 77, 5973–5980, 2011b.
Zwietering, M. H., Jongenburger, I., Rombouts, F. M., and van't Riet, K.: Modeling of the bacterial growth curve, Appl. Environ. Microbiol., 56, 1875–1881, 1990.





