the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
A portable biosensor for simultaneous diagnosis of TNF-α and IL-1β in saliva biomarkers using twin electronic devices
Majid Monajjemi
Fatemeh Mollaamin
Motahareh Dehghandar
Sara Shahriari
Parisa Latifi
Samira Mohammadi
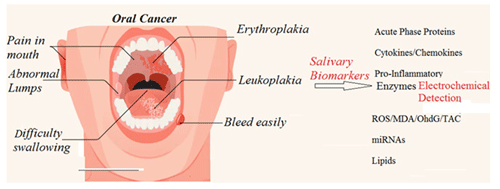
For several types of diseases, such as meningitis or oral cancers, the simultaneous diagnosis and measurement of the tumour necrosis factor α (TNF-α) and interleukin-1 β (IL-1β) immune-modulating biomarkers, with respect to both quality and quantity, are important. For example, although meningitis is generally caused by bacteria or viruses, the differences between viral and bacterial structures can be problematic for medical doctors to distinguish, as laboratory data assay techniques for the two are often similar and can overlap; moreover, in such cases, distinguishing between virial and bacterial structures is especially problematic following the use of antibiotics prior to cerebrospinal fluid testing. In this work, we simultaneously evaluated the precision of both TNF-α and IL-1β for the diagnosis of disease. In this research area, twin electrochemical biosensors have been designed as strong tools for the wide-spectrum assessment of biomarkers, thereby aiding in the screening, diagnosis, and monitoring of pathologies and treatment performance. In this research, we present a sensor platform model that can enable one to detect biomarkers quickly; specifically, this platform can be used to detect TNF-α and IL-1β in saliva. A two-peptide recognition element was created and designed using the phage display technique. This element selectively binds TNF-α and IL-1β to an electronics-based metal–oxide–semiconductor field-effect transistor/electrolyte-gated transistor (MOSFET/GT) bio-detector device and label-free biosensor, allowing for the rapid, simultaneous detection of both biomarkers. These bio-affinity recognition methods have been successfully implemented to realize the experimental twin-model sensor, based on electrolyte-gated transistor (EGT) and semiconductor field-effect transistor (ZnO-SFET) biosensors, to test for these two disease biomarkers, both individually and simultaneously, with high performance. In summary, we developed a sensor platform that can be used for rapid oral cancer signature analysis of biomarkers in multiple bio-fluids of saliva. This system works by arraying metal–oxide–semiconductor field-effect transistors (MOSFETs), with each targeting a biorecognition element (BRE) specific to one of these two important biomarkers. This system can also be extended upon to aid in a wide variety of cancer research applications.
- Article
(2688 KB) - Full-text XML
- BibTeX
- EndNote
1.1 Saliva protein biomarker detection and related diseases
Biomarkers, which are found in several tissues of our biological systems, have been applied in medical diagnostics (Abayomi et al., 2006; Ryu et al., 2015). However, as the application of one biomarker is not sufficient to detect a disease, we focus on the detection of multiple biomarkers in this work, with the aim of diagnosing any type of cancer (Choi et al., 2010; Zadeh et al., 2015). Because the concentration of biomarkers in the human body is usually restricted to a narrow range, we evaluated two types of biosensor models for advanced medical diagnosis: one contained an accurate detection biosensor, whereas the other was a low-cost, portable sensing device (Cunningham and Laing, 2008). Although, nowadays, due to the use of modern systems that combine various technologies, most biomarker sensors have decreased limitations with respect to detection, there are several major questions that need still to be discussed:
-
How do we improve the performance of biomarkers?
-
How do we obtain a suitable signal intensity for specific biomarkers?
-
How can the concentration of biomaterials be converted into changes in the properties of the chemical structure or microstructure of these compounds?
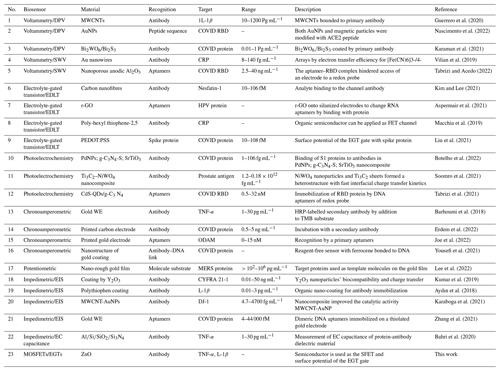
The detection of protein biomarkers in saliva is usually confirmed via enzyme-linked immune sorbent assay (ELISA) techniques (Gug et al., 2019). Based on this method, the protein on the ELISA microtiter plate is fixed by direct adsorption through an antibody that is precoated on the surface. Detection is accomplished by loading an enzyme-labelled antibody and observing its interaction with the substrate, which causes a luminescent signal. Another possible method, known as western blotting, is based on the immunological system and is generally applied in studies in salivary biomarkers (Hsiao et al., 2018). Liquid chromatography–tandem mass spectrometry (LC-MS-MS) is another high-throughput technique for screening the saliva proteome and has been used by researchers such as Hsiao et al. (2018) to screen hundreds of peptides in several protein biomarkers. Moreover, gel electrophoresis can be used for the analysis of around 100 biomarker proteins; however, advanced MS techniques, such as matrix-assisted laser desorption–ionization time-of-flight mass spectrometry (MALDI-ToF MS), are sometimes needed. Despite their broad versatility and quick analysis times, these techniques require an intensive sample pretreatment process prior to any analysis (Hu et al., 2007; Kipping et al., 2021). Raman and nuclear magnetic resonance (NMR; Sarasia et al., 2011; Monajjemi et al., 2008) spectroscopy are much better methods (compared with LC-MS-MS) from the perspective of protein profiling; this is due to the simple sample preparation processes required as well as the fact that Raman scattering is based on the frequency shift interaction of light with proteins in the laser radiation. It is notable that the Raman signals need to be applied via metallic nanostructures before any measurement takes place, for example, when examining biomarker protein salivary cytokines with Gold (Au) nanorods to diagnose bronchial inflammation in asthmatic patients using a surface-enhanced Raman spectroscopy (SERS) technique (Zamora-Mendoza et al., 2019). Silver nanoparticles have also been used, via a SERS technique, to aid in the diagnosis of lung cancer in patients by analysing salivary biomarker proteins (Li et al., 2012). A list of barcodes of various biomarker proteins can be extracted for further detection via signal processing enhancement and the use of SERS techniques (Buchan et al., 2021). Salivary proteins have been related to some localized and systemic diseases. Recent research has reported the importance of salivary C-reactive protein (CRP) as a confirmatory biomarker of acute myocardial infarction (Patel et al., 2015; Bellagambi et al., 2021). Cytokines, which are the main subject of our research work, are largely produced in the oral cavity and are therefore related to oral pathologies (Kaushik et al., 2011; Costa et al., 2010). Interleukins (ILs; including IL-1β and IL-6), tumour necrosis factor α (TNF-α), and matrix metalloproteinases have been identified widely and applied as important biomarkers for the diagnosis of several diseases (Dikova et al., 2021; Asatsuma et al., 2004). Levels of salivary IL-8 have been discovered to cause bowel diseases and muscle diseases (Rathnayake et al., 2013). IL-19 has been confirmed as an indicator of asthma severity as well as being a potential biomarker for therapy of this disease (Wang et al., 2017; Xiao et al., 2016). As complex devices and complicated techniques are not suitable for the fast detection of disease, we have designed and developed a novel twin-biomarker sensor for the rapid testing and detection of salivary markers in this work. For this purpose, we initially began by analysing and evaluating various biosensors, such as electronic sensors (Wei et al., 2013), electrochemical sensors (Gug et al., 2019), fluorescent sensors (Christodoulides et al., 2005), interferometer sensors (Samavati et al., 2021), plasmatic sensors (Guerreiro et al., 2014), absorbance sensors (Park et al., 2012), and quartz crystal microbalance sensors (Arif et al., 2015) (Table 1), with respect to their applications in the detection of protein biomarkers in saliva. We found that the electrochemical sensor was the best compromise between low cost and high analytical performance among the existing sensor platforms. In this paper, we first discuss these two biomarkers comprehensively and then analyse their properties and structures.
1.2 Tumor necrosis factor α (TNF-α) and interleukin-1 β (IL-1β)
TNF compounds are cytokines belonging to the TNF family, which consists of several transmembrane proteins (Mollaamin et al., 2015, 2011), including a homologous TNF subunit (Liu et al., 2020; Gravallese and Monach, 2015). It was the first cytokine to be discovered to be an adipocyte, as secreted by adipose tissue (Sethi and Hotamisligil, 2021). TNF signalling appears via tumour necrosis factor receptor 1 (TNFR1) and TNFR2 (Heir and Stellwagen, 2020; Gough and Myles, 2020); although TNFR1 is expressed in most cells, TNFR2 is primarily restricted to endothelial, epithelial, and subsets of immune cells (Rolski and Błyszczuk, 2020). There are two types of TNF-α, including m-TNF-α and s-TNF-α, with the latter being a soluble form (Qu et al., 2017; Probert, 2015). The main function of TNF is in the regulation of immune cells, although it also acts as an endogenous pyrogen and is able to prevent apoptotic cell death via inflammation and the inhibition tumour genesis.
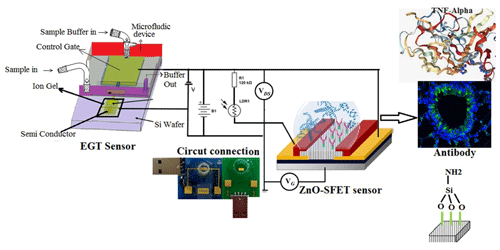
Due to the perfect properties of electrochemical sensor systems with respect to protein biomarker detection, these sensor techniques have primarily been used in the study of salivary biomarkers (Kakino et al., 2018; Chu et al., 2013; Ramadori and Armbrust, 2001). Moreover, due to their high efficiency, portability, low cost, and simple operation, electrochemical sensors are able detect saliva proteins with minimum time, for example, without adding extra electrolytes (Dharmalingam and Yamasandhi, 2018). Thus, this work was based on the electrochemical analyses of saliva samples via the exploitation of the relatively strong ionic strength of saliva. The concept roadmap for the electrochemical detection of protein biomarkers in saliva is depicted in Fig. 1. In the electrochemical immunoassay, the attached biomarkers, with the conjugated antibodies as the specific recognition elements, induce a rapid electrode response via voltage changes, current changes, or impedance variation. As the aptamers can basically recognize the probes for protein biomarkers, the antibodies should preferably be located on the electrodes directly using nanomaterial (Liang et al., 2008). Aptamers with high stability can automatically change their size and structure compared with antibodies (Wandrer et al., 2020; Kanda et al., 2006). On the other hand, nanobodies are unique compounds of bio-receptors with a high interest with respect to binding to the electrochemical proteins' structures in any type of biosensor (Murao et al., 2000; Kakino et al., 2018). Nanobodies are small antibodies with a higher affinity for targets with a higher stability and suitable solubility (due to their lower dimensions) compared with antibodies (Mani et al., 2021). Based on these concepts and properties, we designed an artificial antibody using a molecularly imprinted polymer (MIP) for our electrochemical biosensors.
In this research, specific peptides have been synthesized for TNF-α and 1L-1β with no use of biomolecular probes, and enzymatic reactions involving the hydrolysis of the target and subsequent activation of redox probes have been suggested as novel strategy for TNF-α and IL-6 detection. Moreover, functional chemical groups from several nanomaterial surfaces have been used in order to recognize the target protein in the presence of activating agents. These selected biomolecular probes and functional chemical groups have basically been used to modify the electrode of our sensor. We used a mix of several electrochemical sensors, including amperometric sensors, voltammetric sensors, electrochemical impedance spectroscopy (EIS), and electrochemical capacitance sensors (Torrente-Rodríguez et al., 2016). In potentiometric techniques, the electrochemical potential measured between two electrodes varies in the analyte over one of the electrodes (Pires et al., 2011). Electrolyte-gated transistors (EGTs) are widely used in special protein detection and have a mechanism based on metal–oxide–semiconductor field-effect transistors (MOSFETs; Burtscher et al., 2021; Gualandi et al., 2019). The difference between MOSFET and EGT methods depends on the material type used: the former uses a dielectric material for the gate, whereas the latter uses an electrolyte as the gate.
We have investigated an electronics-based (MOSFET/EGT) label-free biosensor that provides rapid detection of TNF-α and 1L-1β in saliva. The biorecognition elements (BREs) that bind TNF-α and 1L-1β were selected using a phage display technique (detailed in Sect. 3.1.1) and confirmed via both binding kinetics and docking software and molecular mechanic modelling. We used this powerful tool to isolate specific peptide/antibody binders against the target of interest. Consequently, using this technique, we are able to determine the tumour antigens based on protein–DNA interactions using DNA libraries with randomized segments.
3.1 Phage display
3.1.1 Phage display library against TNF-α and 1L-1β
An enzyme-linked immune-sorbent assay (ELISA) microtiter plate was coated with 80 µ of 0.2 mg mL−1 human TNF-α (Gen-Script Corporation, Piscataway, USA). It was also coated with 100 µ of 0.5 mg mL−1 human interleukin-β (1L-1β) (Gen-Script Corporation, Piscataway, USA). A total of 0.2 M NaHCO3 buffer was added to each mixture at pH = 7.4. The plates then underwent a 20 h incubation at 5 °C in a humid chamber. After 2 h incubation at 5 °C in a humid chamber, the solution was mixed slowly (over 4 h) with serum albumin (6 mg mL−1), 0.2 M NaHCO3 (pH = 7.4), and NaN3 = 0.05 %) and washed with tris-buffered saline (TBS), 60 mM tris-HCl, and 200 mM NaCl using the Phage Display Peptide Library containing 2 × 1010 phages in 150 µ of TBS-T 0.2 % (PhD-C7C, New England Biolabs Inc., Westburg B. X., Leusden, the Netherlands). After rinsing, TNF-α- and 1L-1β-bound phages were eluted with 0.5 M glycine-HCl buffer (pH = 2.5) and then complemented with 0.1 % bovine serum albumin (BSA); following this stage, they were neutralized with 1 M tris-HCl buffer (pH = 8.1). Amplified phages were obtained at 5 °C in a specific solution containing 25 % polyethylene glycol and 2.0 M NaCl. The phage mixture was added to a TBS buffer (100 mM tris-HCl and 200 mM NaCl at pH = 7.4). Escherichia coli was grown on a plate containing isopropyl-β-D-thiogalactoside (IPTG; ICN Biomedical Inc., Germany) and 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside (Xgal; Sigma-Aldrich, Germany). To determine the sequences of selected phage clones, the Sanger method was used, in which nucleotide DNA chains terminate (Sambrook et al., 1989).
3.1.2 Selection of the peptide sequences through docking simulation
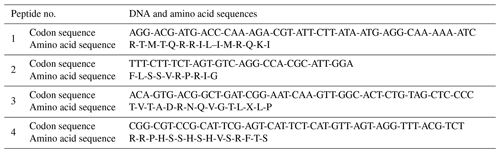
Peptide sequences were selected via a docking simulation of 7kpb.pdb as well as 1TNF.pdb and 9ilb.pdb crystallography for both the TNF-α and 1L-1β structures (as shown in Fig. 2). In addition, several monomer subunits were used for further description. The potential binding sites were calculated and plotted using Acsite software. Docking was performed with the iGEMDOCK algorithms. In the first step, the interactive maps of the docked molecules were created using LIGPLOT (Wallace et al., 1995). The docked minimum energies were then analysed in order to find suitable places where any type of peptide could attach with higher affinity to the TNFα. Three of the peptides docked exhibited suitable binding energies, and docking was repeated with an increased stringency using a reduced cavity size as a receptor. The reverse complement of the DNA insert was obtained and then translated to give the amino acid product of the DNA insert (Monajjemi et al., 2014, 2010b) (Table 2). This resulted in one matching peptide that was again modelled with TNFα (Protein Data Bank, PDB: 1TNF) and 1L-1β as a template using the MODELLER software (Sali et al., 1995; Wang et al., 2002).
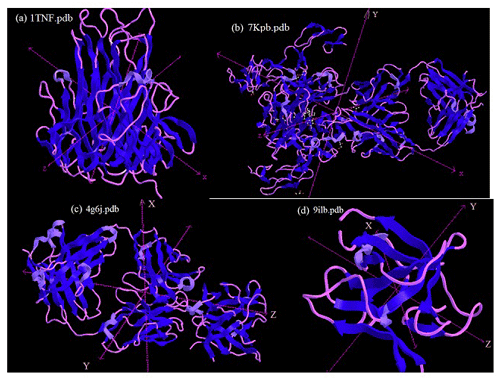
Figure 2(a) The structure of tumour necrosis factor α (TNFα) and its implications for receptor binding; (b) the human TNFα TNFR1 complex bound to a conformational selective antibody; (c) the crystal structure of human IL-β; (d) human interleukin-β (1L-1β). This figure was plotted using the Chem3D software.
Candidate peptides were obtained via phage display, and two sequences of interest, RTMTQRILIMRQKI for TNF-α binding and FLSSVRPRIG for 1L-1β binding, referred to as Peptide 1 and Peptide 2, respectively, were identified. To accurately determine the binding kinetics of the Peptide1-TNF-α and Peptide2-1L-1β complexes, several concentrations of TNF-α-binding and 1L-1β-binding peptides were injected over a streptavidin surface plasmon resonance (SPR) chip using a Biacore T200 system. Kinetic fits indicate that these two peptides bind with higher affinity to TNF-α and 1L-1β biomarkers (Table 3). The dissociation constant KD was calculated using the following formula: KD = .
Table 3Equilibrium dissociation constants for peptide1-TNF-α and peptide2-1L-1β complexes containing docking energies.

* VDW: van der Waals interaction.
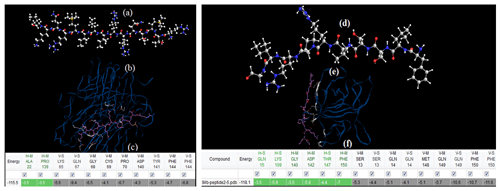
Figure 3(a) Optimization and configurations of Peptide 1 including the RTMTQRILIMRQKI amino acid sequence, using CHARMM software. (b) Peptide 1–TNFα docked using iGEMDOCK software. (c) Amino acids with related energies during interaction with TNF-α. (d) Optimization and configurations of Peptide 2 including the FLSSVRPRIG amino acid sequence, using CHARMM software. (e) Peptide 2–1L-1β docked using iGEMDOCK software. (f) Amino acids with related energies during interaction with 1L-1β. This figure was plotted using docking (iGEMDOCK) software.
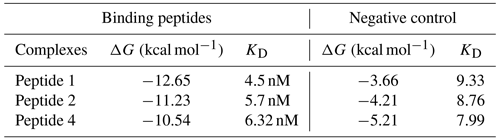
The two aforementioned peptides with the addition of a negative control peptide (NCP) created by scrambled binding with sequence TVTADRNQVGTLXLP (Table 3) were used in this study. In total, we generated an ensemble configuration for Peptide1-TNF-α, Peptide-2-1L-1β, and NCP using replica exchange molecular dynamics based on Sugita and Okamoto (1999). The docking of peptides binding with TNF-α and 1L-1β was accomplished with two steps: (1) using the PatchDock package and (2) performing rotational and translational positioning to generate a set of possible complexes. We observed which two peptides attached to the proper region of the TNF-α and 1L-1β biomarkers via a combination of electrostatics, hydrogen bonding, and van der Waals interactions; in the proposed structure of the complex, residue Ala to Phe and residue Gln to Phe, for Peptide 1 (Table 3 and Fig. 3) and Peptide 2, respectively, are located on the side of the proper region of the TNF-α and 1L-1β biomarkers, permitting correct attachment to the (MOSFETs/EGTs) surfaces. NCP also binds to the biomarkers for any further comparison, although at a much lower level compared with the two biomarkers (Table 4).
Initially, the docking configurations for TNF-α-Peptide-1 and NCP were scored with the FireDock energy function (Andrusier et al., 2007). This function considers the atomic contact energy (ACE), van der Waals interactions, partial electrostatics, hydrogen and disulfide bonds, p-stacking and aliphatic interactions, and additional terms (Andrusier et al., 2008). We calculated the Gibbs energies of complexes using molecular dynamics (MDs) simulations with the CHARMM software. The absolute binding free energy was calculated using the molecular mechanics Poisson–Boltzmann surface area (MM-PBSA) methodology (Kollman et al., 2000): ΔG (binding) = . To prepare a system for MM-PBSA calculations, the complexes were solvated in a rectangular box of TIP3P water molecules.
3.1.3 RTMTQRILIMRQKI and FLSSVRPRIG peptide synthesis
Based on the docking simulation, the two proper conformations of the RTMTQRILIMRQKI and FLSSVRPRIG peptides with the complex system should be sensitized for use in the bioelectric sensor model. Solid-phase peptide synthesis was performed by converting amino acids to hydroxybenzotriazole-activated esters by treatment with hydroxybenzotriazole (HOBT) and diisopropylcarbodiimide in dimethylformamide (DMF). The subsequent reactions were accomplished in DMF. After synthesis, side-chain-protecting groups were removed, and the peptides were cleaved from the support resin with trifluoroacetic acid. The inhibition of the cytotoxic effect of TNF-α and 1L-1β by the phages or the RTMTQRILIMRQKI and FLSSVRPRIG peptides was assayed using the saliva proteins. Briefly, cells were plated into microtiter plates (NUNC, Roskilde, Germany) using Dulbecco's modified eagle medium (DMEM) and cultured overnight at 300 K in the presence of 10 % CO2. After removing the supernatant, both 1L-1β and TNF-α were incubated separately with various concentrations of each phage clone or peptide in complete DMEM supplemented with 3 mg mL−1 actinomycin D-mannitol for 3 h at 300 K in 10 % CO2; thereafter, the TNF-α-RTMTQRILIMRQKI and 1L-1β-FLSSVRPRIG from phage mixtures were added to the two separated cell cultures for overnight incubation at 300 K in 10 % CO2. After removing the supernatant, absorbance was measured at 550 nm using a Dynatech MR 5000 microplate reader (Chantilly, VA, USA). Each concentration of phage of peptide-TNF-α and peptide-1L-1β mixtures was tested in triplicate.
3.1.4 MOSFET/EGT sensor development and testing
In our model sensor, MOSFETs were fabricated based on a zinc oxide field-effect transistor (ZnO-SFET). This offers various advantages: first, ZnO is suitable for pulsed laser deposition under ambient conditions, thereby lending itself to use in a low-cost, portable sensing device that can be employed everywhere and at any time; second, a ZnO-SFET can be fabricated using a variety of materials for any type of packaging, such as flexible and lightweight plastics; third, as ZnO can be deposited in a suitable situation, it automatically results in a vertically aligned nanostructure, yielding a semiconductor with a wide surface area, which is perfect for ZnO-SFET sensors. Although various high-performance ZnO-SFETs have recently been investigated to increase the speed of electronic models (Bayraktaroglu et al., 2009; Hagen et al., 2011), our twin-model sensor based on electrolyte-gated transistors (EGTs) and semiconductor field-effect transistors (MOSFETs) is a unique device that can be widely used to detect several special proteins simultaneously (Fig. 4).
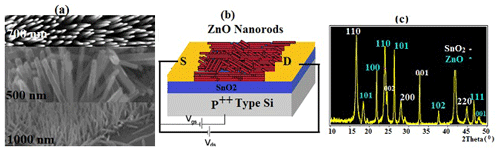
3.1.5 Fabrication of nanoscale ZnO semiconductors
Patterned semiconductor nanostructures were integrated with electrical contacts and isolated using a passivation layer (Aroonyadet et al., 2015; Fortunato et al., 2012). During combination with a patterning process, a ZnO-SFET was fabricated using top-down vapour-phase thin-film deposition. Basically, ZnO-FET devices have good electrical properties with field-effect mobility > 15 cm2 V−1 s−1 and an on/off current ratio from 105 to 108; therefore, they are compatible with high sensing performance (Liu et al., 2018, 2016) (Fig. 5). Figure 5c exhibits the X-ray diffraction (XRD) pattern of the fabricated SnO2–ZnO composite heterojunction material. The heterojunction between SnO2 and ZnO increased the performance of a ZnO-SFET gated by SnO2 between the source (S) and drain (D).
In this work, a range of bio-recognition approaches, including enzyme–substrate interactions, antibody–antigen interactions, and nucleic acid hybridization, were studied as biosensors for a specific molecular target (Table 1). Among these biomarkers, we focused on several compounds on rows 1, 13, 19, and 22 of Table 1 that were related to two types of antibodies for two targets, including 1L-1β and TNF-α, using conjugate biomarkers in saliva. These bio-affinity recognition methods have been successfully implemented to realize an experimental twin-model sensor based on electrolyte-gated transistor (EGT) and semiconductor field-effect transistor (ZnO-SFETs) biosensors for testing for these two disease biomarkers, either individually or simultaneously, and have shown high performance. Using a docking simulation to examine the effect of different solvents and temperatures on the stability of a single-walled carbon nanotube (Khaleghian et al., 2011; Monajjemi et al., 2010a), we developed a nano-ZnO-SFET biosensing device for the saliva biomarker of our molecular bio-receptor with high and specific binding affinities for the target of TNF-α on the surface of the SFET. In addition, a sensor based on electrolyte-gated transistors (EGTs) was also suitable for 1L-1β and showed high performance. ZnO-based SFETs were, therefore, generally functionalized first with a chemical agent to enable covalent immobilization of the specific bio-receptors on their surface (Fig. 4). In this regard, we used mercaptopropyltrimethoxysilane (MPTMS), applied as organosilane families that covalently and easily bind to ZnO (Fleischhaker et al., 2010). Silanization was carried out in either the vapour or liquid phase using a 90 % 10 % () mixture of ethanol water and the Liu method (Liu et al., 2013). The amine functional groups of 3-aminopropyltriethoxysilane (APTES) can be bound to glutaraldehyde and, consequently, are able to create a covalent bond with conjugate bio-receptors containing the TNF-α antibody. Meanwhile MPTMS can create a covalent bond with conjugate bio-receptors containing the 1L-1β antibody. After fixing the unreacted groups, such as the CHO–SiO2 group, which are generally blocked, the surface is passive toward reducing non-specific adsorption events (Fig. 6; Heiney et al., 2000; Zheng et al., 2005).
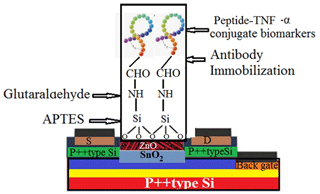
Figure 6The SnO2–ZnO thin-film-based biosensor's surface functionalization process involves silanization with APTES, reaction with glutaraldehyde, and covalent antibody conjugation.
In parallel, 1L-1β was injected through the sample port of the EGT sensor (Fig. 5), and L-1β-bound phages containing a buffer at pH = 2.5 with 0.1 % BSA were also added. Due to this fact, the ZnO anchor sequence preferentially bound to the semiconductor over any other surface on the SFET array. Figure 7a shows the area in which the morphology of the SiO2 surface is 100 % flat; however, the ZnO surface morphology changes due to the attachment of the bifunctional RTMTQRILIMRQKI peptide to the semiconductor (Fig. 7b). The functionalized ZnO of the ZnO-SFET results in high efficiency in both the section containing the gate-to-source voltage (VGS) and that containing the source-to-drain voltage (VSD) (Fig. 7c).
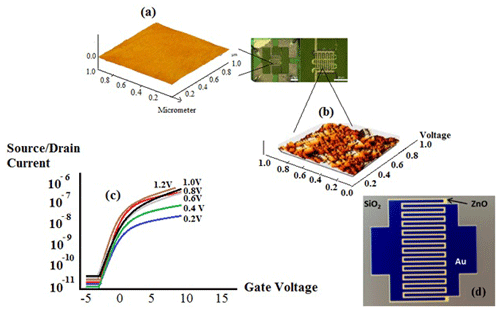
Figure 7(a) Optical micrograph of the interdigital electrode ZnO-SFET structure. (b) An atomic force micrograph of the functionalized peptide SFET containing a mix of SiO2 morphology and ZnO morphology. (c) The peptide SFET dry-state efficiency with modulation of the VGS and VSD. (d) The binding and dry position efficiency of the ZnO-SFET.
In Fig. 7d, the active ZnO electrode, comprised of ZnO nanorods, acted to keep each SFET electrically isolated from its adjacent area. The diced SFET chips (13.5 mm × 4.5 mm) were incubated in the peptide solution for 10 min and were then washed and dried several times. We utilized a general technique, based on Hermanson (2010), to bind BREs to surfaces via covalent linker chemistry. The anchoring peptide domain is based on the sequence developed by Tomczak et al. (2009), which binds to ZnO. The RTMTQRILIMRQKI peptide was added to the TNF-α, and the placement of the alanine end of the Peptide-TNF-α sequence to the linker was specifically chosen based on computational modelling methods that showed the importance of the amino acids on the phenylalanine (Fig. 3). A quick sensor response was obtained by presenting the peptide SFET to several certain concentrations of the TNF-α target while also monitoring the source/drain current (ISD), at fixed constant VGS and VSD values. We rehydrated the peptide SFET in solution using liquid sensing to ensure the effective binding of TNF-α. For this purpose, 25 µL of sterile water (Gibco ultrapure DNase/RNase-free water, 18.1 MΩ) was inserted into the devices, and we monitored the ISD until it was fixed. The sensor was exposed to a solution containing 15 nM TNF-α several times (each time for 2 min), and the source/drain current was monitored each time. By enhancing concentrations of TNF-α, a concomitant increase in current appeared; however, the magnitude of the current increase began to decrease at higher concentrations due to the saturation of the peptide binding sites for three solvents (Fig. 8a). By measuring each quick static response in each of the concentrations, the difference in current change can be determined for the target exposure. The quick nature of the TNF-α detection in sterile water is shown in Fig. 8b (for three solvents), where a current increase is immediately seen following the application of a 9 fM droplet of TNF-α. In addition, the solvent effect can be seen in these plots (using three colours: green, blue, and red). In the green plot, pure ethanol has been used, which has a dielectric constant of around 24.5, and a minimum current can be seen at time = 40 s. In the blue plot, using 10 % water (blue) causes the minimum position to move to around time = 50 s, whereas using 90 % water (red plot) decreases this amount to time = 22. This dielectric effect also helps us to calibrate our sensor based on a solvent and the dielectric effect.
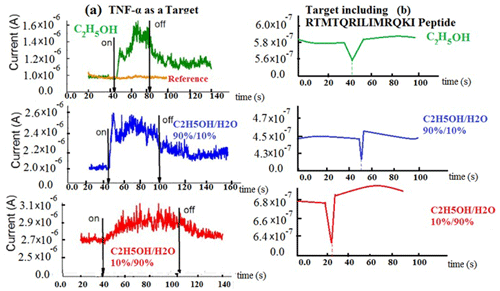
Figure 8Increasing concentrations of TNF-α versus time in three solvents (a) without peptide and (b) containing peptide.
Before the filtration process and injection of the liquid (Fig. 4), the sensitivity to Peptide-1-SFET and Peptide-2-EGT in a saliva matrix was seen in several scanning ranges, from 0.2 up to 1.2 µM, according to the elution time in the absorbance chromatogram. Consequently, we selected 0.5 µM for TNF-α and 0.8 µM for 1L-1β, due to perfect adsorption at these concentrations (Fig. 9a). After filtration, when spiked into a 20 µL drop of human saliva, Peptide-1-SFET and Peptide-2-EGT exhibited significant responses to 20 fM (2 × 10–14) for TNF-α and to 15 fM for 1L-1β (Fig. 9b). As can be seen from Fig. 9a, the absorbance has a maximum of 3.2 min according to the elution time. In addition, the source (S) and drain (D) current in Fig. 9b exhibits 15 and 20 fM sensitivities for Peptide-2-EGT and Peptide-1-SFET, respectively.
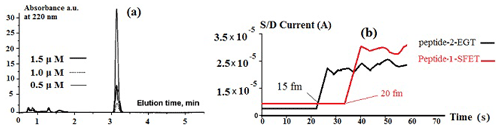
Figure 9(a) Sensitivity of Peptide-1-SFET in a saliva matrix with scanning ranges of 0.2–1.2 µM. (b) A rapid significant response to 20 fM (2 × 10–14) is found for TNF-α in water, whereas a significant response to 15 fM is observed for 1L-1β in water, for VGS = 6 V and VSD = 0.4 V.
In Fig. 10, which displays a drain current (ID) versus drain-to-source voltage (VSD) plot, the curve at different gate-to-source voltage (VGS) values for this biosensor has been plotted at different voltages. As can be seen, by increasing the different voltage from −6 V towards +6 V, the slope of the drain current changes (red, green, and blue curves). This changing voltage is an important issue for our fabricated sensor with respect to detecting TNF-α and 1L-1β.
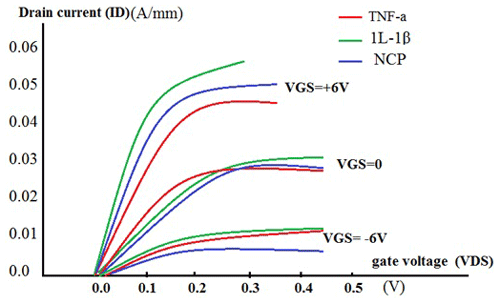
Figure 10A drain current (ID) versus drain-to-source voltage (VSD) curve at different gate-to-source voltage (VGS) values.
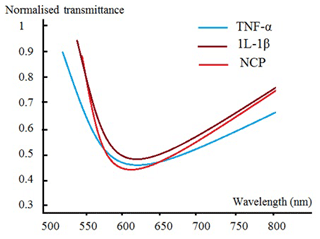
Moreover, for a more rigorous validation of our methodology and to confirm the accuracy of this biosensor, we used some other sensing platforms, such as electrochemical biomarker sensors (EBSs) and an SPR-based biomarker sensor. As the mechanism of the SPR sensor is based on electrode charge transfer after the specific binding of biomarkers, the change in material properties or electronic states of sensors is caused by antigen–antibody specific binding, which produces different electrical signals at different concentrations (Oh et al., 2004; Lee et al., 2004; Guan et al., 2005). SPR is an optical physical phenomenon whereby the surface of a metal generates plasma waves when polarized light reflects onto a metal film at a certain angle. When SPR is applied to the biomarker detection field, the biomarker-specific binding changes the refractive index of the plasma resonant material and causes a change in the resonant angle of the SPR that can be measured to detect the concentration of biomarker. SPR-based biomarker sensors always have ultrahigh sensitivity and resolution. Figure 11 exhibits our fabricated low-cost SPR biosensor with a plastic optical-fibre waveguide for the detection of oral cancer in our system containing TNF-α, 1L-1β, and NCP.
It is notable that our fabricated sensor offers numerous advantages due to its cheap price, accurate measurement, and portability; moreover, it is suitable for use in many applications, such as medical diagnosis, drug predication, drug delivery (Khalili Hadad et al., 2011; Ghalandari et al., 2011) and integration in electronic instruments to examine biological molecules' behaviours via a sensing electrode for the detection of target issues. As a result, this sensor, which is applied to detect oral cancer, helps us get one step closer to clinical application. This sensor is a dynamic platform for biosensing cancer biomarkers using a twin-model electrochemical biosensor in the screening, diagnosis, and monitoring of pathologies and treatment performance. The main novelty of this work is an in-depth study of the capability of a simple sensor platform model that enables one to quickly detect TNF-α and IL-1β in saliva biomarkers.
Although many biomarker sensors have been developed in recent years, the performance of commercial sensors applied in the clinical field has not improved significantly. Considering modern medical diagnosis requirements (which are mentioned in Sect. 1), the development and optimization of practical commercial sensors is of paramount importance compared with that of laboratory products. We believe that, with continued advancements in materials science and microfabrication technologies, some practical biomarker sensors could break through existing bottlenecks, optimize performance, and achieve breakthroughs with respect to human health in the future.
The field of application for this sensor is rapid detection in biological fluids, such as saliva and serum, in which sub-nanomole detection is needed. We exhibited a sensor platform that can be used for the rapid analysis of an oral cancer signature using biomarkers in multiple bio-fluids of saliva. This system works by arraying a MOSFET/EGT system, with each transistor targeting a BRE specific to one of two important biomarkers. This system can also be extended and applied to a wide variety of cancer research applications. One of the major challenges in the development of a twin bioelectronic sensor is the progress of biosensing unit and signal transducer techniques, which should be considered in detail. In summary, as the development of modern materials and microfabrication technologies advance, biomarker sensors and platforms will not only break through current challenges but will also exhibit tremendous potential for the early diagnosis of cancer as well as individualized health monitoring (Monajjemi et al., 2011). Continuous monitoring using this sensor is a promising solution for employment in clinical practice and in the home setting. Using this sensor, patients' diseases can be monitored during hospitalization to detect clinical deterioration. A modified early-warning score (MEWS), a scoring system incorporating all intermittent measurements and other observations, is often used to facilitate the detection of clinical deterioration in nursing wards. Although implementation of this technology for continuous monitoring in clinical practice is a complex process, this can be affected by technical, social, and organizational factors.
| ACE2 | Human angiotensin-converting enzyme 2 |
| CdS-QDs | Cadmium sulfide quantum dots |
| COVID | Coronavirus disease |
| CRP | C-reactive protein |
| DPV | Differential pulse voltammetry |
| EGT | Electrolyte-gated transistor |
| EIS | Electrochemical impedance spectroscopy |
| EC | Electrochemical |
| EDLT | Electrical double-layer transistor |
| FET | Field-effect transistor |
| g-C3N4 | Graphite carbon nitride |
| HPV | Human papillomavirus |
| HRP | Horseradish peroxidase |
| MERS | Middle East respiratory syndrome coronavirus |
| MWCNT | Multi-walled carbon nanotube |
| MWCNT-AuNP | Multi-walled carbon nanotube Au nanoparticle |
| ODAM | Human odontogenic ameloblast-associated protein |
| PdNP | Pd nanoparticle |
| PEDOT | Poly-3,4-ethylenedioxythiophene |
| PSS | Polystyrene sulfonate |
| RBD | Receptor-binding domain |
| r-GO | Reduced graphene oxide |
| SWV | Square wave voltammetry |
| TMB | Tetramethylbenzidine |
The CHARMM code is available from https://www.academiccharmm.org/ (CHARMM, 2024); HyperChem is available from http://www.hypercubeusa.com/ (Hypercube, Inc., 2024); RasMol V2.7.5 is available from http://www.rasmol.org/software/rasmol/INSTALL.html (RasMol, 2024); iGEMDOCK is available from https://www.tbi.org.tw/tools/download.php (TBI, 2024); and ChemDraw Professional and Chem3D are available from https://guides.library.stanford.edu/scilib-software/chemdraw (Robin Li and Melissa Ma Science Library, 2024).
Data are not publicly accessible, as a patent application is currently pending. Once the patent is granted, the data may become available from the authors upon reasonable request.
MM designed the research method and verified and accomplished the molecular docking situation and related methodology; they are also the corresponding author and take responsibility for the paper; MM, MD, and SM designed, measured, and drew the electronics, including images, graphs, and experimental activities. PL and SS performed and checked the biological analysis and abbreviation; MM and FM wrote the manuscript; FM revised the manuscript and checked dictation and grammar.
The contact author has declared that none of the authors has any competing interests.
Publisher’s note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
The authors thank Islamic Azad University for providing the computer equipment and software.
This paper was edited by Michele Penza and reviewed by two anonymous referees.
Abayomi, L. A., Terry, L. A., White, S. F., and Warner, P. J.: Development of a disposable pyruvate biosensor to determine pungency in onions (Allium cepa L.), Biosens. Bioelectron., 21, 2176–2179, https://doi.org/10.1016/j.bios.2005.10.024, 2006.
Arif, S., Qudsia, S., Urooj, S., Chaudry, N., Arshad, A., and Andleeb, S.: Blueprint of quartz crystal microbalance biosensor for early detection of breast cancer through salivary autoantibodies against ATP6AP1, Biosens. Bioelectron., 65, 62–70, https://doi.org/10.1016/j.bios.2014.09.088, 2015.
Andrusier, N., Nussinov, R., and Wolfson, H. J.: FireDock: Fast Interaction Refinement in Molecular Docking, Proteins, 69, 139–159, https://doi.org/10.1002/prot.21495, 2007.
Andrusier, N., Mashiach, E., Nussinov, R., and Wolfson, H. J.: Principles of flexible protein–protein docking, Proteins, 73, 271–289, https://doi.org/10.1002/prot.22170, 2008.
Aroonyadet, N., Wang, X., Song, Y., Chen, H., Cote, R. J., Thompson, M. E., Datar, R. H., and Zhou, C.: Highly Scalable, Uniform, and Sensitive Biosensors Based on Top-Down Indium Oxide Nanoribbons and Electronic Enzyme-Linked Immunosorbent Assay, Nano Lett., 15, 1943–1951, https://doi.org/10.1021/nl5047889, 2015.
Asatsuma, M., Ito, S., Watanabe, M., Takeishi, H., Nomura, S., Wada, Y., Nakano, M., Gejyo, F., and Igarashi, A.: Matrix metalloproteinase 9 (MMP-9) gene polymorphism, Clin. Chim. Acta, 345, 99–104, https://doi.org/10.1016/j.cccn.2004.03.006, 2004.
Aspermair, P., Mishyn, V., Bintinger, J., Happy, H., Bagga, K., Subramanian, P., Knoll, W., Boukherroub, R., and Szunerits, S.: Reduced graphene oxide–based field effect transistors for the detection of E7 protein of human papillomavirus in saliva, Anal. Bioanal. Chem., 413, 779–787, https://doi.org/10.1007/s00216-020-02879-z, 2021.
Aydın, M., Aydın, E. B., and Sezgintürk, M. K.: A highly selective electrochemical immunosensor based on conductive carbon black and star PGMA polymer composite material for IL-8 biomarker detection in human serum and saliva, Biosens. Bioelectron., 117, 720–728, https://doi.org/10.1016/j.bios.2018.07.010, 2018.
Bahri, M., A. Baraket, N., Zine, M. B., Ali, J., Bausells, A., and Errachid, T.: Capacitance electrochemical biosensor based on silicon nitride transducer for TNF-α cytokine detection in artificial human saliva: Heart failure (HF), Talanta, 209, 120501, https://doi.org/10.1016/j.talanta.2019.120501, 2020.
Barhoumi, L., Baraket, A., Bellagambi, F. G., Karanasiou, G. S., Ali, M. B., Fotiadis, D. I., Bausells, J., Zine, N., Sigaud, M., and Errachid, A.: A novel chronoamperometric immunosensor for rapid detection of TNF-α in human saliva, Sensor. Actuat. B-Chem., 266, 477–484, https://doi.org/10.1016/j.snb.2018.03.135, 2018.
Bayraktaroglu, B., Leedy, K., and Neidhard, R.: Highfrequency ZnO thin-film transistors on Si substrates, IEEE Electr. Device L., 30, 946–948, 2009.
Bellagambi, F. G., Petersen, C., Salvo, P., Ghimenti, S., Franzini, M., Biagini, D., Hangouët, M., Trivella, M. G., Di Francesco, F., Paolicchi, A., Errachid, A., Fuoco, R., and Lomonaco, T.: Determination and stability of N-terminal pro-brain natriuretic peptide in saliva samples for monitoring heart failure, Sci. Rep., 11, 13088, https://doi.org/10.1038/s41598-021-92488-2, 2021.
Botelho, C. N., Falcão, S. S., Soares, R.-E. P., Pereira, S. R., de Menezes, A. S., Kubota, L. T., Damos, F. S., and Luz, R. C. S.: Evaluation of a photoelectrochemical platform based on strontium titanate, sulfur doped carbon nitride and palladium nanoparticles for detection of SARS-CoV-2 spike glycoprotein S1, Biosensors and Bioelectronics: X, 11, 100167, https://doi.org/10.1016/j.biosx.2022.100167, 2022.
Buchan, E., Kelleher, L., Clancy, M., Rickard, J. J. S., and Oppenheimer, P. G.: Spectroscopic molecular-fingerprint profiling of saliva, Anal. Chim. Acta, 1185, 339074, https://doi.org/10.1016/j.aca.2021.339074, 2021.
Burtscher, B., Urbina, P. A. M., Diacci, C., Borghi, S., Pinti, M., Cossarizza, A., Salvarani, C., Berggren, M., Biscarini, F., Simon, D. T., and Bortolotti, C. A.: Sensing Inflammation Biomarkers with Electrolyte-Gated Organic Electronic Transistors, Adv. Healthc. Mater., 10, 2100955, https://doi.org/10.1002/adhm.202100955, 2021.
CHARMM (Chemistry at HARvard Macromolecular Mechanics): https://www.academiccharmm.org/, last access: 22 November 2024.
Choi, Y. E., Kwak, J. W., and Park, J. W.: Nanotechnology for early cancer detection, Sensors, 10, 428–455, https://doi.org/10.3390/s100100428, 2010.
Christodoulides, N., Mohanty, S., Miller, C. S., Langub, M. C., Floriano, P. N., Dharshan, P., Ali, M., Bernard, F., Romanovicz, B. D., Anslyn, E., Fox, P. C., and McDevitt, J. T.: Application of microchip assay system for the measurement of C-reactive protein in human saliva, Lab Chip, 5, 261–269, 2005.
Chu, P. S., Nakamoto, N., Ebinuma, H., Usui, S., Saeki, K., and Matsumoto, A.: C-C motif chemokine receptor 9 positive macrophages activate hepatic stellate cells and promote liver fibrosis in mice, Hepatology, 58, 337–350, https://doi.org/10.1002/hep.26351, 2013.
Costa, P. P., Trevisan, G. L., Macedo, G. O., Palioto, D. B., Souza, S. L. S., Grisi, M. F. M., Novaes Jr., A. B., and Taba Jr., M.: Salivary interleukin-6, matrix metalloproteinase-8, and osteoprotegerin in patients with periodontitis and diabetes, J. Periodontol., 81, 384, https://doi.org/10.1902/jop.2009.090510, 2010.
Cunningham, B. T. and Laing, L. G.: Advantages and application of label-free detection assays in drug screening, Expert Opin. Drug Dis., 3, 891–901, https://doi.org/10.1517/17460441.3.8.891, 2008.
Dikova, V., Jantus-Lewintre, E., and Bagan, J.: Potential Non-Invasive Biomarkers for Early Diagnosis of Oral Squamous Cell Carcinoma, J. Clin. Med., 10, 1658, https://doi.org/10.3390/jcm10081658, 2021.
Dharmalingam, M. and Yamasandhi, P. G.: Nonalcoholic Fatty Liver Disease and Type 2 Diabetes Mellitus, Indian Journal of Endocrinology and Metabolism, 22, 421–428, https://doi.org/10.4103/ijem.IJEM_585_17, 2018.
Erdem, A., Senturk, H., Yildiz, E., and Maral, M.: Amperometric immunosensor developed for sensitive detection of SARS-CoV-2 spike S1 protein in combined with portable device, Talanta, 244, 123422, https://doi.org/10.1016/j.talanta.2022.123422, 2022.
Fleischhaker, F., Wloka, V., and Hennig, I.: ZnO based field-effect transistors (FETs): Solution-processable at low temperatures on flexible substrates, J. Mater. Chem., 20, 6622–6625, 2010.
Fortunato, E., Barquinha, P., and Martins, R.: Oxide semiconductor thin-film transistors: A review of recent advances, Adv. Mater., 24, 2945–2986, 2012.
Ghalandari, B., Monajjemi, M., and Mollaamin, F.: Theoretical investigation of carbon nanotube binding to DNA in view of drug delivery, J. Comput. Theor. Nanos., 8, 1212–1219, https://doi.org/10.1166/jctn.2011.1801, 2011.
Gough, P. and Myles, I. A.: Tumor Necrosis Factor Receptors: Pleiotropic Signaling Complexes and Their Differential Effects, Front. Immunol., 11, 585880, https://doi.org/10.3389/fimmu.2020.585880, 2020.
Gravallese, E. M. and Monach, P. A.: 94 - The rheumatoid joint: Synovitis and tissue destruction, Rheumatology (Sixth Edition), 1, 768–784, https://doi.org/10.1016/B978-0-323-09138-1.00094-2, 2015.
Gualandi, I., Tessarolo, M., Mariani, F., Tonelli, D., Fraboni, B., and Scavetta, E., Organic Electrochemical Transistors as Versatile Analytical Potentiometric Sensors, Front. Bioeng. Biotechnol., 7, 354, https://doi.org/10.3389/fbioe.2019.00354, 2019.
Guan, W.-J., Li, Y., Chen, Y. Q., Zhang, X. B., and Hu, G. Q.: Glucose biosensor based on multi-wall carbon nanotubes and screen printed carbon electrodes, Biosens. Bioelectron., 21, 508–512, 2005.
Guerreiro, J. R. L., Frederiksen, M., Bochenkov, V. E., De Freitas, V., and Sales, M. G. F.: Multifunctional biosensor based on localized surface plasmon resonance for monitoring small molecule–protein interaction, ACS Nano, 8, 7958–7967, https://doi.org/10.1021/nn501962y, 2014.
Guerrero, S., Agüí, L., Yáñez-Sedeño, P., and Pingarrón, J. M.: Design of electrochemical immunosensors using electro-click chemistry. Application to the detection of IL-1β cytokine in saliva, Bioelectrochemistry, 133, 107484, https://doi.org/10.1016/j.bioelechem.2020.107484, 2020.
Gug, I. T., Tertis, M., Hosu, O., and Cristea, C.: Salivary biomarkers detection: Analytical and immunological methods overview, TrAC-Trend. Anal. Chem., 113, 301–316, 2019.
Hagen, J., Kim, S., Bayraktaroglu, B., Leedy, K., Chavez, J., Kelley-Loughnane, N., Naik, R., and Stone, M.: Biofunctionalized Zinc Oxide Field Effect Transistors for Selective Sensing of Riboflavin with Current Modulation, Sensors, 11, 6645–6655, https://doi.org/10.3390/s110706645, 2011.
Heiney, P. A., Grüneberg, K., Fang, J., Dulcey, C., and Shashidhar, R.: Structure and Growth of Chromophore-Functionalized (3-Aminopropyl)triethoxysilane Self-Assembled on Silicon, Langmuir, 16, 2651–2657, 2000.
Heir, R. and Stellwagen, D.: TNF-Mediated Homeostatic Synaptic Plasticity: From in vitro to in vivo Models, Front. Cell. Neurosci., 14, 565841, https://doi.org/10.3389/fncel.2020.565841, 2021.
Hermanson, G.: Bioconjugate Techniques, Elsevier, New, York, 2nd edn., eBook ISBN: 9780080568720, 2010.
Hsiao, Y.-C., Chu, L. J., Chen, Y.-T., Chi, L.-M., Chien, K.-Y., Chiang, W.-F., Chang, Y.-T., Chen, S.-F., Wang, W.-S., Chuang, Y.-N., Lin, S.-Y., Chien, C.-Y., Chang, K.-P., Chang, Y.-S., and Yu, J.-S.: Variability Assessment of 90 Salivary Proteins in Intraday and Interday Samples from Healthy Donors by Multiple Reaction Monitoring-Mass Spectrometry, Proteomic. Clin. Appl., 12, 1700039, https://doi.org/10.1002/prca.201700039, 2018.
Hu, S., Loo, J. A., and Wong, D. T.: Human saliva proteome analysis, Ann. NY Acad. Sci., 1098, 323, https://doi.org/10.1196/annals.1384.015, 2007.
Hypercube, Inc.: HyperChem, http://www.hypercubeusa.com/, last access: 22 November 2024.
Joe, C., Lee, B. H., Kim, S. H., Kod, Y., and Gu, M. B.: Aptamer duo-based portable electrochemical biosensors for early diagnosis of periodontal disease, Biosens. Bioelectron., 199, 113884, https://doi.org/10.1016/j.bios.2021.113884, 2022.
Kakino, S., Ohki, T., Nakayama, H., Yuan, X., Otabe, S., Hashinaga, T., Wada, N., Kurita, Y., Tanaka, K., Hara, K., Soejima, E., Tajiri, Y., and Yamada, K.: Pivotal Role of TNF-α in the Development and Progression of Nonalcoholic Fatty Liver Disease in a Murine Model, Horm. Metab. Res., 50, 80–87, https://doi.org/10.1055/s-0043-118666, 2018.
Kanda, H., Tateya, S., Tamori, Y., Kotani, K., Hiasa, K., Kitazawa, R., Kitazawa, S., Miyachi, H., Maeda, S., Egashira, K., and Kasuga, M.: MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity, The Journal of Clinical Investigation, 116, 1494–1505, https://doi.org/10.1172/JCI26498, 2006.
Karaboga, M. N. S. and Sezgintürk, M. K.: A nano-composite based regenerative neuro biosensor sensitive to Parkinsonism-associated protein DJ-1/Park7 in cerebrospinal fluid and saliva, Bioelectrochemistry, 138, 107734, https://doi.org/10.1016/j.bioelechem.2020.107734, 2021.
Karaman, C., Yola, B. B., Karaman, O., Atar, N., Polat, I., and Yola, M. L.: Sensitive sandwich-type electrochemical SARS-CoV2 nucleocapsid protein immunosensor, Microchim. Acta, 188, 425, https://doi.org/10.1007/s00604-021-05092-6, 2021.
Kaushik, R., Yeltiwar, R. K., and Pushpanshu, K.: Salivary interleukin-1β levels in patients with chronic periodontitis before and after periodontal phase I therapy and healthy controls: a case-control study, J. Periodontol., 82, 1353–1359, https://doi.org/10.1902/jop.2011.100472, 2011.
Khaleghian, M., Zahmatkesh, M., Mollaamin, F., and Monajjemi, M.: Investigation of solvent effects on armchair single-walled carbon nanotubes: A QM/MD study, Fuller. Nanotub. Car. N., 19, 251–261, https://doi.org/10.1080/15363831003721757, 2011.
Khalili Hadad, B., Mollaamin, F., and Monajjemi, M.: Biophysical chemistry of macrocycles for drug delivery: A theoretical study, Russ. Chem. Bull., 60, 238–241, https://doi.org/10.1007/s11172-011-0039-5, 2011.
Kim, S. G. and Lee, J. S. J.: Multiscale pore contained carbon nanofiber-based field-effect transistor biosensors for nesfatin-1 detection, Mater. Chem. B, 9, 6076–6083, 2021.
Kipping, M., Tänzler, D., and Sinz, A.: A rapid and reliable liquid chromatography/mass spectrometry method for SARS-CoV-2 analysis from gargle solutions and saliva, Anal. Bioanal. Chem, 413, 6503, https://doi.org/10.1007/s00216-021-03614-y, 2021.
Kollman, P. A., Massova, I., Reyes, C., Kuhn, B., Huo, S., Chong, L., Lee, M., Lee, T., Duan, Y., Wang, W., Donini, O., Cieplak, P., Srinivasan, J., Case, D. A., and Cheatham, T. E.: III. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models, Acc. Chem. Res., 33, 889–897, 2000.
Kumar, S., Panwar, S., Augustine, S., and Malhotra, B. D.: Biofunctionalized Nanostructured Yttria Modified Non-Invasive Impedometric Biosensor for Efficient Detection of Oral Cancer, Nanomaterials, 9, 1190, https://doi.org/10.3390/nano9091190, 2019.
Lee, J., Yoon, K. H., Hwang, K. S., Park, J., Ahn, S., and Kim, T. S.: Label free novel electrical detection using micromachined PZT monolithic thin fi lm cantilever for the detection of Creactive protein, Biosens. Bioelectron., 20, 269–275, 2004.
Lee, W.-I., A. Subramanian, S., Mueller, K., Levon, C.-Y., and Nam, M. H.: Rafailovich, Potentiometric biosensors based on molecular-imprinted self-assembled monolayer films for rapid detection of influenza a virus and SARS-CoV-2 spike protein, ACS Appl. Nano Mater, 5, 5045, https://doi.org/10.1021/acsanm.2c00068, 2022.
Li, X., Yang, T., and Lin, J.: Spectral analysis of human saliva for detection of lung cancer using surface-enhanced Raman spectroscopy, J. Biomed. Opt., 17, 037003, https://doi.org/10.1117/1.JBO.17.3.037003, 2012.
Liang, H., Yin, B., Zhang, H., Zhang, S., Zeng, Q., and Wang, J.: Blockade of tumor necrosis factor (TNF) receptor type 1-mediated TNF-α signaling protected Wistar rats from diet-induced obesity and insulin resistance, Endocrinology, 149, 2943–2951, https://doi.org/10.1210/en.2007-0978, 2008.
Liu, C. Y., Tam, S. S., Huang, Y., Dubé, P. E., Alhosh, R., Girish, N., Punit, P., Nataneli, S., Li, F., Bender, J. M., Washington, M. K., and Polk, D. B.: TNF Receptor 1 Promotes Early-Life Immunity and Protects against Colitis in Mice, Cell Reports, 33, 108275, https://doi.org/10.1016/j.celrep.2020.108275, 2020.
Liu, H., Yang, A., Song, J., Wang, N., Lam, P., Li, Y., Law, H. K.-W., and Yan, F.: Ultrafast, sensitive, and portable detection of COVID-19 IgG using flexible organic electrochemical transistors, Sci. Adv., 7, 8387, https://doi.org/10.1126/sciadv.abg8387, 2021.
Liu, Q., Liu, Y., Wu, F., Cao, X., Li, Z., Alharbi, M., Abbas, A.N., Amer, M.R., and Zhou, C.: Highly Sensitive and Wearable In2O3 Nanoribbon Transistor Biosensors with Integrated On-Chip Gate for Glucose Monitoring in Body Fluids, ACS Nano, 12, 1170–1178, 2018.
Liu, Q., Aroonyadet, N., Song, Y., Wang, X., Cao, X., Liu, Y., Cong, S., Wu, F., Thompson, M. E., and Zhou, C.: Highly Sensitive and Quick Detection of Acute Myocardial Infarction Biomarkers Using In2O3 Nanoribbon Biosensors Fabricated Using Shadow Masks, ACS Nano, 10, 10117–10125, 2016.
Liu, Y., Li, Y., Li, X.-M., and He, T.: Kinetics of (3-Aminopropyl)triethoxylsilane (APTES) Silanization of Superparamagnetic Iron Oxide Nanoparticles, Langmuir, 29, 15275–15282, 2013.
Macchia, E., Manoli, K., Holzer, B., Franco, C. D., Picca, R. A., Cioffi, N., Scamarcio, G., Palazzo, G., and Torsi, L.: Selective single-molecule analytical detection of C-reactive protein in saliva with an organic transistor, Anal. Bioanal. Chem, 411, 4899, https://doi.org/10.1007/s00216-019-01778-2, 2019.
Mani, V., Beduk, T., Khushaim, W., Ceylan, A. E., Timur, S., Wolfbeis, O. S., and Salama, K. N.: Electrochemical sensors targeting salivary biomarkers: A comprehensive review TrAC, Trends Anal. Chem., 135, 116164, https://doi.org/10.1016/j.trac.2020.116164, 2021.
Mollaamin, F., Najafi, F., Khaleghian, M., Hadad, B. K., and Monajjemi, M.: Theoretical study of different solvents and temperatures effects on single-walled carbon nanotube and temozolomide drug: A QM/MM study, Fuller. Nanotub. Car. N., 19, 653–667, https://doi.org/10.1080/1536383X.2010.504956, 2011.
Mollaamin, F., Ilkhani, A., Sakhaei, N., Bonsakhteh, B., Faridchehr, A., Tohidi, S., and Monajjemi, M.: Thermodynamic and solvent effect on dynamic structures of nano bilayer-cell membrane: Hydrogen bonding study, J. Comput. Theor. Nanos., 12, 3148–3154, https://doi.org/10.1166/jctn.2015.4092, 2015.
Monajjemi, M., Afsharnezhad, S., Jaafari, M. R., Mirdamadi, S., Mollaamin, F., and Monajemi, H.: Investigation of energy and NMR isotropic shift on the internal rotation Barrier of Θ4 dihedral angle of the DLPC: A GIAO study, Chemistry, 17, 55–69, 2008.
Monajjemi, M., Khaleghian, M., Tadayonpour, N., and Mollaamin, F.: The effect of different solvents and temperatures on stability of single-walled carbon nanotube: A QM/MD study, Int. J. Nanosci., 9, 517–529, https://doi.org/10.1142/S0219581X10007071, 2010a.
Monajjemi, M., Noei, M., and Mollaamin, F.: Design of fMet-tRNA and calculation of its bonding properties by quantum mechanics, Nucleos. Nucleot. Nucl., 29, 676–683, https://doi.org/10.1080/15257771003781642, 2010b.
Monajjemi, M., Khosravi, M., Honarparvar, B., and Mollaamin, F. : Substituent and solvent effects on the structural bioactivity and anticancer characteristic of catechin as a bioactive constituent of green tea, Int. J. Quantum Chem., 111, 2771–2777, https://doi.org/10.1002/qua.22612, 2011.
Monajjemi, M., Yamola, H., and Mollaamin, F.: Study of bio-nano interaction outlook of amino acids on single-walled carbon nanotubes, Fuller. Nanotub. Car. N., 22, 595–603, https://doi.org/10.1080/1536383X.2012.702163, 2014.
Murao, K., Ohyama, T., Imachi, H., Ishida, T., Cao, W. M., and Namihira, H.: TNF-alpha stimulation of MCP-1 expression is mediated by the Akt/PKB signal transduction pathway in vascular endothelial cells, Biochem. Bioph. Res. Co., 276, 791–796, https://doi.org/10.1006/bbrc.2000.3497, 2000.
Nascimento, E. D., Fonseca, W. T., de Oliveira, T. R., de Correia, C. R. S. T. B., Faça, V. M., de Morais, B. P., Silvestrini, V. C., Pott-Junior, H., Teixeira, F. R., and Faria, R. C.: COVID-19 diagnosis by SARS-CoV-2 Spike protein detection in saliva using an ultrasensitive magneto-assay based on disposable electrochemical sensor, Sensor. Actuat. B-Chem., 353, 131128, https://doi.org/10.1016/j.snb.2021.131128, 2022.
Oh, B. K., Lee, W., Kim, Y. K., Lee, W. H., and Choi, J.-W.: Surface plasmon resonance immunosensor using self-assembled protein G for the detection of Salmonella paratyphi, J. Biotechnol., 111, 1–8, 2004.
Park, J., Sunkara, V., Kim, T.-H., Hwang, H., and Cho, Y.-K.: Lab-on-a-disc for fully integrated multiplex immunoassays, Anal. Chem., 84, 2133, https://doi.org/10.1021/ac203163u, 2012.
Patel, N., Belcher, J., Thorpe, G., Forsyth, N. R., and Spiteri, M. A.: Measurement of C-reactive protein, procalcitonin and neutrophil elastase in saliva of COPD patients and healthy controls: correlation to self-reported wellbeing parameters, Resp. Res., 16, 62, https://doi.org/10.1186/s12931-015-0219-1, 2015.
Pires, N. M. M., Dong, T., Yang, Z., Høivik, N., and Zhao, X.: A mediator embedded micro-immunosensing unit for electrochemical detection on viruses within physiological saline media, J. Micromech. Microeng., 21, 115031, https://doi.org/10.1088/0960-1317/21/11/115031, 2011.
Probert, L.: TNF and its receptors in the CNS: The essential, the desirable and the deleterious effects, Neuroscience, 302, 2–22, https://doi.org/10.1016/j.neuroscience.2015.06.038, 2015.
Qu, Y., Zhao, G., and Li, H.: Forward and Reverse Signaling Mediated by Transmembrane Tumor Necrosis Factor-Alpha and TNF Receptor 2: Potential Roles in an Immunosuppressive Tumor Microenvironment, Front. Immunol., 8, 1675, https://doi.org/10.3389/fimmu.2017.01675, 2017.
Ramadori, G. and Armbrust, T.: Cytokines in the liver, Eur. J. Gastroen. Hepat., 13, 777–784, https://doi.org/10.1097/00042737-200107000-00004, 2001.
RasMol: Installation Instructions, RasMol 2.7.5, http://www.rasmol.org/software/rasmol/INSTALL.html, last access: 22 November 2024.
Rathnayake, N., Åkerman, S., Klinge, B., Lundegren, N., Jansson, H., Tryselius, Y., Sorsa, T., and Gustafsson, A.: Salivary Biomarkers for Detection of Systemic Diseases, PLoS One, 8, e61356, https://doi.org/10.1371/journal.pone.0061356, 2013.
Robin Li and Melissa Ma Science Library: General Overview of ChemDraw and Signals Notebook, Stanford Libraries, https://guides.library.stanford.edu/scilib-software/chemdraw, last access: 22 November 2024.
Rolski, F. and Błyszczuk, P.: Complexity of TNF-α Signaling in Heart Disease, J. Clin. Med., 9, 3267, https://doi.org/10.3390/jcm9103267, 2020.
Ryu, J., Lee, E., Lee, K., and Jang, J.: A graphene quantum dots based fluorescent sensor for anthrax biomarker detection and its size dependence, J. Mater. Chem. B, 3, 4865–4870, 2015.
Sali, A., Potterton, L., Yuan, F., van Vlijmen, H., and Karplus, M.: Evaluation of comparative protein modeling by MODELLER, Proteins, 23, 318–326, 1995.
Samavati, A., Samavati, Z., Velashjerdi, M., Ismail, A. F., Othman, M. H. D., Eisaabadi, B. G., Abdullah, M. S., Bolurian, M., and Bolurian, M.: Sustainable and fast saliva-based COVID-19 virus diagnosis kit using a novel GO-decorated Au/FBG sensor, Chem. Eng. J., 420, 127655, https://doi.org/10.1016/j.cej.2020.127655, 2021.
Sambrook, J., Fritsch, E. F., and Maniatis, T.: Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, New York, NY, USA, ISBN (Hardback): 978-0-87969-309-1, 1989.
Sarasia, E. M., Afsharnezhad, S., Honarparvar, B., Mollaamin, F., and Monajjemi, M.: Theoretical study of solvent effect on NMR shielding tensors of luciferin derivatives, Phys. Chem. Liq., 49, 561–571, https://doi.org/10.1080/00319101003698992, 2011.
Sethi, J. K. and Hotamisligil, G. S.: Metabolic Messengers: tumour necrosis factor, Nature Metabolism, 3, 1302–1312, https://doi.org/10.1038/s42255-021-00470-z, 2021.
Soomro, R. A., Jawaid, S., Zhang, P., Han, X., Hallam, K. R., Karakuş, S., Kilislioğlu, A., Xu, B., and Willander, M.: NiWO4-induced partial oxidation of MXene for photo-electrochemical detection of prostate-specific antigen, Sensor. Actuat. B Chem., 328, 129074, https://doi.org/10.1016/j.snb.2020.129074, 2021.
Sugita, Y. and Okamoto, Y.: Replica-exchange molecular dynamics method for protein folding, Chem. Phys. Lett., 314, 141–151, 1999.
Tabrizi, M. A. and Acedo, P.: An electrochemical membrane-based aptasensor for detection of severe acute respiratory syndrome coronavirus-2 receptor-binding domain, Appl. Surf. Sci., 598, 153867, https://doi.org/10.1016/j.apsusc.2022.153867, 2022.
Tabrizi, M. A., Nazari, L., and Acedo, P.: A photo-electrochemical aptasensor for the determination of severe acute respiratory syndrome coronavirus 2 receptor-binding domain by using graphitic carbon nitride-cadmium sulfide quantum dots nanocomposite, Sensor. Actuat. B-Chem., 345, 130377, https://doi.org/10.1016/j.snb.2021.130377, 2021.
TBI (Taiwan Bioinformatics Institute): iGEMDOCK Generic Evolutionary Method for molecular DOCKing, https://www.tbi.org.tw/tools/download.php, last access: 22 November 2024.
Tomczak, M., Gupta, M., Drummy, L., Rozenzhak, S., and Naik, R.: Morphological control and assembly of zinc oxide using a biotemplate, Acta Biomater., 5, 876–882, 2009.
Torrente-Rodríguez, R. M., Campuzano, S., Ruiz-Valdepeñas Montiel, V., Gamella, M., and Pingarrón, J. M.: Electrochemical bioplatforms for the simultaneous determination of interleukin (IL)-8 mRNA and IL-8 protein oral cancer biomarkers in raw saliva, Biosens. Bioelectron., 77, 543, https://doi.org/10.1016/j.bios.2015.10.016, 2016.
Vilian, A. T. E., Kim, W., Park, B., Oh, S. Y., Kim, T., Huh, Y. S., Hwangbo, C. K., and Han, Y.-K.: Efficient electrn-mediated electrochemical biosensor of gold wire for the rapid detection of C-reactive protein: A predictive strategy for heart failure, Biosens. Bioelectron., 142, 111549, https://doi.org/10.1016/j.bios.2019.111549, 2019.
Wallace, A. C., Laskowski, R. A., and Tornton, J. M.: LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions, Protein Eng., 8, 127–134, 1995.
Wandrer, F., Liebig, S., Marhenke, S., Vogel, A., John, K., Manns, M. P., Teufel, A., Itzel, T., Longerich, T., Maier, O., Fischer, R., Kontermann, R. E., Pfizenmaier, K., Schulze-Osthoff, K., and Bantel, H.: TNF-Receptor-1 inhibition reduces liver steatosis, hepatocellular injury and fibrosis in NAFLD mice, Cell Death Dis., 11, 212, https://doi.org/10.1038/s41419-020-2411-6, 2020.
Wang, X., Kaczor-Urbanowicz, K. E., and Wong, D. T. W.: Salivary Biomarkers in Cancer Detection, Med. Oncol., 34, 7, https://doi.org/10.1007/s12032-016-0863-4, 2017.
Wang, R., Lai, L., and Wang, S.: Further development and validation of empirical scoring functions for structure-based binding affinity prediction, J. Comput. Aid. Mol. Des., 16, 11–26, 2002.
Wei, F., Yang, J., and Wong, D. T. W.: Detection of exosomal biomarker by electric field-induced release and measurement (EFIRM), Biosens. Bioelectron., 44, 115, https://doi.org/10.1016/j.bios.2012.12.046, 2013.
Xiao, H., Zhang, Y., Kim, Y., Kim, S., Kim, J. J., Kim, K. M., Yoshizawa, J., Fan, L.-Y., Cao, C.-X., and Wong, D. T. W.: Differential Proteomic Analysis of Human Saliva using Tandem Mass Tags Quantification for Gastric Cancer Detection, Sci. Rep., 6, 22165, https://doi.org/10.1038/srep22165, 2016.
Yousefi, H., Mahmud, A., Chang, D., Das, J., Gomis, S., Chen, J. B., Wang, H., Been, T., Yip, L., Coomes, E., Li, Z., Mubareka, S., McGeer, A., Christie, N., Gray-Owen, S., Cochrane, A., Rini, J. M., Sargent, E. H., and Kelley, S. O.: Detection of SARS-CoV-2 Viral Particles Using Direct, Reagent-Free Electrochemical Sensing, J. Am. Chem. Soc., 143, 1722–1727, https://doi.org/10.1021/jacs.0c10810, 2021.
Zadeh, M. A. A., Lari, H., Kharghanian, L., Balali, E., Khadivi, R., Yahyaei, H., Mollaamin, F., and Monajjemi, M.: Density functional theory study and anti-cancer properties of shyshaq plant: In view point of nano biotechnology, J. Comput. Theor. Nanos., 12, 4358–4367, https://doi.org/10.1166/jctn.2015.4366, 2015.
Zamora-Mendoza, B. N., Espinosa-Tanguma, R., Ramírez-Elías, M. G., Cabrera-Alonso, R., Montero-Moran, G., Portales-Pérez, D., Rosales-Romo, J. A., Gonzalez, J. F., and Gonzalez, C.: Surface-enhanced raman spectroscopy: A noninvasive alternative procedure for early detection in childhood asthma biomarkers in saliva, Photodiagn. Photodyn., 27, 85–91, 2019.
Zhang, Z., Pandey, R., Li, J., Gu, J., White, D., Stacey, H. D., Ang, J. C., Steinberg, C.-J., Capretta, A., Filipe, C. D. M., Mossman, K., Balion, C., Miller, M. S., Salena, B. J., Yamamura, D., Soleymani, L., Brennan, J. D., and Li, Y.: High-Affinity Dimeric Aptamers Enable the Rapid Electrochemical Detection of Wild-Type and B.1.1.7 SARS-CoV-2 in Unprocessed Saliva, Angew. Chem. Int. Edit., 60, 24266–24274, https://doi.org/10.1002/anie.202110819, 2021.
Zheng, G., Patolsky, F., Cui, Y., Wang, W. U., and Lieber, C. M.: Multiplexed electrical detection of cancer markers with nanowire sensor arrays, Nat. Biotechnol., 23, 1294–1301, 2005.
- Abstract
- Introduction
- Electrochemical sensing systems
- Experimental and molecular modelling details
- Results and discussion
- Future perspectives
- Conclusion
- Appendix A: Abbreviations
- Code availability
- Data availability
- Author contributions
- Competing interests
- Disclaimer
- Acknowledgements
- Review statement
- References
- Abstract
- Introduction
- Electrochemical sensing systems
- Experimental and molecular modelling details
- Results and discussion
- Future perspectives
- Conclusion
- Appendix A: Abbreviations
- Code availability
- Data availability
- Author contributions
- Competing interests
- Disclaimer
- Acknowledgements
- Review statement
- References